Genome-wide identification and evolution of the SAP gene family in sunflower (Helianthus annuus L.) and expression analysis under salt and drought stress
- PMID: 39099650
- PMCID: PMC11296301
- DOI: 10.7717/peerj.17808
Genome-wide identification and evolution of the SAP gene family in sunflower (Helianthus annuus L.) and expression analysis under salt and drought stress
Abstract
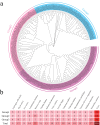
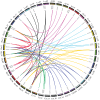
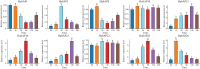
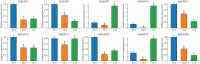
Stress-associated proteins (SAPs) are known to play an important role in plant responses to abiotic stresses. This study systematically identified members of the sunflower SAP gene family using sunflower genome data. The genes of the sunflower SAP gene family were analyzed using bioinformatic methods, and gene expression was assessed through fluorescence quantification (qRT-PCR) under salt and drought stress. A comprehensive analysis was also performed on the number, structure, collinearity, and phylogeny of seven Compositae species and eight other plant SAP gene families. The sunflower genome was found to have 27 SAP genes, distributed across 14 chromosomes. The evolutionary analysis revealed that the SAP family genes could be divided into three subgroups. Notably, the annuus variety exhibited amplification of the SAP gene for Group 3. Among the Compositae species, C. morifolium demonstrated the highest number of collinearity gene pairs and the closest distance on the phylogenetic tree, suggesting relative conservation in the evolutionary process. An analysis of gene structure revealed that Group 1 exhibited the most complex gene structure, while the majority of HaSAP genes in Group 2 and Group 3 lacked introns. The promoter analysis revealed the presence of cis-acting elements related to ABA, indicating their involvement in stress responses. The expression analysis indicated the potential involvement of 10 genes (HaSAP1, HaSAP3, HaSAP8, HaSAP10, HaSAP15, HaSAP16, HaSAP21, HaSAP22, HaSAP23, and HaSAP26) in sunflower salt tolerance. The expression of these 10 genes were then examined under salt and drought stress using qRT-PCR, and the tissue-specific expression patterns of these 10 genes were also analyzed. HaSAP1, HaSAP21, and HaSAP23 exhibited consistent expression patterns under both salt and drought stress, indicating these genes play a role in both salt tolerance and drought resistance in sunflower. The findings of this study highlight the significant contribution of the SAP gene family to salt tolerance and drought resistance in sunflower.
Keywords: Evolutionary analysis; SAP; Sunflower; qRT-PCR.
© 2024 Zhang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Genome-wide identification and expression analysis of ZF-HD family in sunflower (Helianthus annuus L.) under drought and salt stresses.BMC Plant Biol. 2025 Feb 3;25(1):140. doi: 10.1186/s12870-025-06139-z. BMC Plant Biol. 2025. PMID: 39894816 Free PMC article.
-
Genome-wide identification of the TIFY gene family in Helianthus annuus and expression analysis in response to drought and salt stresses.Sci Rep. 2025 Apr 30;15(1):15138. doi: 10.1038/s41598-025-99315-y. Sci Rep. 2025. PMID: 40307306 Free PMC article.
-
Genome-wide identification and characterization of protein phosphatase 2C (PP2C) gene family in sunflower (Helianthus annuus L.) and their expression profiles in response to multiple abiotic stresses.PLoS One. 2024 Mar 20;19(3):e0298543. doi: 10.1371/journal.pone.0298543. eCollection 2024. PLoS One. 2024. PMID: 38507444 Free PMC article.
-
Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.).Gene. 2024 Jun 5;910:148336. doi: 10.1016/j.gene.2024.148336. Epub 2024 Mar 4. Gene. 2024. PMID: 38447680 Review.
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
Cited by
-
Identification and Analysis of Stress-Associated Protein (SAP) Transcription Factor Family Members in Pinus massoniana.Plants (Basel). 2025 May 23;14(11):1592. doi: 10.3390/plants14111592. Plants (Basel). 2025. PMID: 40508267 Free PMC article.
-
Comprehensive identification of GASA genes in sunflower and expression profiling in response to drought.BMC Genomics. 2024 Oct 14;25(1):954. doi: 10.1186/s12864-024-10860-8. BMC Genomics. 2024. PMID: 39402437 Free PMC article.
References
-
- Abdullah-Zawawi MR, Ahmad-Nizammuddin NF, Govender N, Harun S, Mohd-Assaad N, Mohamed-Hussein ZA. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Scientific Reports. 2021;11(1):19678. doi: 10.1038/s41598-021-99206-y. - DOI - PMC - PubMed
-
- Asad MAU, Zakari SA, Zhao Q, Zhou L, Ye Y, Cheng F. Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: premature leaf senescence in plants. International Journal of Molecular Sciences. 2019;20(2):256. doi: 10.3390/ijms20020256. - DOI - PMC - PubMed
-
- Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, Legrand L, Gill N, Kane NC, Bowers JE, Hubner S, Bellec A, Bérard A, Bergès H, Blanchet N, Boniface MC, Brunel D, Catrice O, Chaidir N, Claudel C, Donnadieu C, Faraut T, Fievet G, Helmstetter N, King M, Knapp SJ, Lai Z, Le Paslier MC, Lippi Y, Lorenzon L, Mandel JR, Marage G, Marchand G, Marquand E, Bret-Mestries E, Morien E, Nambeesan S, Nguyen T, Pegot-Espagnet P, Pouilly N, Raftis F, Sallet E, Schiex T, Thomas J, Vandecasteele C, Varès D, Vear F, Vautrin S, Crespi M, Mangin B, Burke JM, Salse J, Muños S, Vincourt P, Rieseberg LH, Langlade NB. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546(7656):148–152. doi: 10.1038/nature22380. - DOI - PubMed
-
- Baidyussen A, Aldammas M, Kurishbayev A, Myrzabaeva M, Zhubatkanov A, Sereda G, Porkhun R, Sereda S, Jatayev S, Langridge P, Schramm C, Jenkins CLD, Soole KL, Shavrukov Y. Identification, gene expression and genetic polymorphism of zinc finger A20/AN1 stress-associated genes, HvSAP, in salt stressed barley from Kazakhstan. BMC Plant Biology. 2020;20(Suppl 1):156. doi: 10.1186/s12870-020-02332-4. - DOI - PMC - PubMed
-
- Billah SA, Khan NZ, Ali W, Aasim M, Usman M, Alezzawi MA, Ullah H. Genome-wide in silico identification and characterization of the stress associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in potato (Solanum tuberosum L.) PLOS ONE. 2022;17(8):e0273416. doi: 10.1371/journal.pone.0273416. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

