Unlocking rivers' hidden diversity and ecological status using DNA metabarcoding in Northwest Spain
- PMID: 39100209
- PMCID: PMC11294579
- DOI: 10.1002/ece3.70110
Unlocking rivers' hidden diversity and ecological status using DNA metabarcoding in Northwest Spain
Abstract
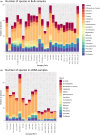
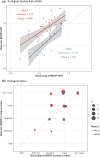
Rivers are crucial ecosystems supporting biodiversity and human well-being, yet they face increasing degradation globally. Traditional river biomonitoring methods based on morphological identification of macroinvertebrates present challenges in terms of taxonomic resolution and scalability. This study explores the application of DNA metabarcoding analysis in both bulk and environmental DNA (eDNA) samples for comprehensive assessment of macrozoobenthic biodiversity, detection of invasive and endangered species, and evaluation of river ecological status in northwestern Spain. DNA metabarcoding of homogenized bulk samples and water eDNA revealed a mean of 100 and 87 macrozoobenthos species per sample respectively. However, the specific composition was significantly different with only 27.3% of the total species being shared. It was not possible to identify all the OTUs to species level; only 17.43% and 49.4% of the OTUs generated could be identified to species level in the bulk and eDNA samples, respectively. Additionally, a total of 11 exotic species (two first records for the Iberian Peninsula and another three first records for Asturias region) and one endangered species were detected by molecular tools. Molecular methods showed significant correlations with morphological identification for EQR values (Ecological Quality Ratio) of IBMWP index, yet differences in inferred river ecological status were noted, with bulk samples tending to indicate higher status. Overall, DNA metabarcoding offers a promising approach for river biomonitoring, providing insights into biodiversity, invasive species, and ecological status within a single analysis. Further optimization and intercalibration are required for its implementation in routine biomonitoring programmes, but its scalability and multi-tasking capabilities position it as a valuable tool for integrated monitoring of river ecosystems.
Keywords: IBMWP; bulk samples; eDNA; ecological status; exotic species; macroinvertebrates.
© 2024 The Author(s). Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Taxonomic accuracy and complementarity between bulk and eDNA metabarcoding provides an alternative to morphology for biological assessment of freshwater macroinvertebrates.Sci Total Environ. 2024 Jul 20;935:173243. doi: 10.1016/j.scitotenv.2024.173243. Epub 2024 May 16. Sci Total Environ. 2024. PMID: 38761946
-
Modeling the ecological status response of rivers to multiple stressors using machine learning: A comparison of environmental DNA metabarcoding and morphological data.Water Res. 2020 Sep 15;183:116004. doi: 10.1016/j.watres.2020.116004. Epub 2020 Jun 15. Water Res. 2020. PMID: 32622231
-
Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method.Sci Total Environ. 2022 Feb 1;806(Pt 2):150587. doi: 10.1016/j.scitotenv.2021.150587. Epub 2021 Sep 25. Sci Total Environ. 2022. PMID: 34582852
-
Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods.Sci Total Environ. 2022 Apr 20;818:151783. doi: 10.1016/j.scitotenv.2021.151783. Epub 2021 Nov 19. Sci Total Environ. 2022. PMID: 34801504 Review.
-
Stochastic process is main factor to affect plateau river fish community assembly.Environ Res. 2024 Aug 1;254:119083. doi: 10.1016/j.envres.2024.119083. Epub 2024 May 11. Environ Res. 2024. PMID: 38735377 Review.
Cited by
-
A Study on the Community and Ecological Characteristics of Benthic Invertebrates in the Ulungu River, Xinjiang, via eDNA Metabarcoding and Morphological Methods.Biology (Basel). 2025 Apr 12;14(4):410. doi: 10.3390/biology14040410. Biology (Basel). 2025. PMID: 40282275 Free PMC article.
References
-
- Alekseev, V. R. (2021). Confusing invader: Acanthocyclops americanus (Copepoda: Cyclopoida) and its biological, anthropogenic and climate‐dependent mechanisms of rapid distribution in Eurasia. Water (Switzerland), 13(10). 10.3390/w13101423 - DOI
-
- Allan, J. D. , Castillo, M. M. , & Capps, K. A. (2021). Stream ecology. Springer International Publishing.
-
- Anderson, L. , & Leite, R. (2012). Mitochondrial pseudogenes in insect DNA barcoding: Differing points of view on the same issue. Biota Neotropica, 12(3), 301–308.
-
- Apothéloz‐Perret‐Gentil, L. , Bouchez, A. , Cordier, T. , Cordonier, A. , Guéguen, J. , Rimet, F. , Vasselon, V. , & Pawlowski, J. (2021). Monitoring the ecological status of rivers with diatom eDNA metabarcoding: A comparison of taxonomic markers and analytical approaches for the inference of a molecular diatom index. Molecular Ecology, 30(13), 2959–2968. 10.1111/mec.15646 - DOI - PMC - PubMed
-
- Barbour, M. T. , Faulkner, C. , & Gerritsen, J. (1998). Rapid bioassessment protocols for use in streams and wadeable rivers: Periphyton, benthic macroinvertebrates, and fish (2nd ed.). U.S. Environmental Protection Agency. http://www.epa.gov/OWOW/monitoring/techmon
LinkOut - more resources
Full Text Sources

