Human Orthohantavirus disease prevalence and genotype distribution in the U.S., 2008-2020: a retrospective observational study
- PMID: 39100240
- PMCID: PMC11296052
- DOI: 10.1016/j.lana.2024.100836
Human Orthohantavirus disease prevalence and genotype distribution in the U.S., 2008-2020: a retrospective observational study
Abstract
Background: In the United States (U.S.), hantavirus pulmonary syndrome (HPS) and non-HPS hantavirus infection are nationally notifiable diseases. Criteria for identifying human cases are based on clinical symptoms (HPS or non-HPS) and acute diagnostic results (IgM+, rising IgG+ titers, RT-PCR+, or immunohistochemistry (IHC)+). Here we provide an overview of diagnostic testing and summarize human Hantavirus disease occurrence and genotype distribution in the U.S. from 2008 to 2020.
Methods: Epidemiological data from the national hantavirus registry was merged with laboratory diagnostic testing results performed at the CDC. Residual hantavirus-positive specimens were sequenced, and the available epidemiological and genetic data sets were linked to conduct a genomic epidemiological study of hantavirus disease in the U.S.
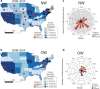
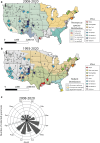
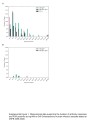
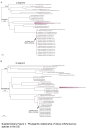
Findings: From 1993 to 2020, 833 human hantavirus cases have been identified, and from 2008 to 2020, 335 human cases have occurred. Among New World (NW) hantavirus cases detected at the CDC diagnostic laboratory (representing 29.2% of total cases), most (85.0%) were detected during acute disease, however, some convalescent cases were detected in states not traditionally associated with hantavirus infections (Connecticut, Missouri, New Jersey, Pennsylvania, Tennessee, and Vermont). From 1993 to 2020, 94.9% (745/785) of U.S. hantaviruses cases were detected west of the Mississippi with 45.7% (359/785) in the Four Corners region of the U.S. From 2008 to 2020, 67.7% of NW hantavirus cases were detected between the months of March and August. Sequencing of RT-PCR-positive cases demonstrates a geographic separation of Orthohantavirus sinnombreense species [Sin Nombre virus (SNV), New York virus, and Monongahela virus]; however, there is a large gap in viral sequence data from the Northwestern and Central U.S. Finally, these data indicate that commercial IgM assays are not concordant with CDC-developed assays, and that "concordant positive" (i.e., commercial IgM+ and CDC IgM+ results) specimens exhibit clinical characteristics of hantavirus disease.
Interpretation: Hantaviral disease is broadly distributed in the contiguous U.S, viral variants are localised to specific geographic regions, and hantaviral disease infrequently detected in most Southeastern states. Discordant results between two diagnostic detection methods highlight the need for an improved standardised testing plan in the U.S. Hantavirus surveillance and detection will continue to improve with clearly defined, systematic reporting methods, as well as explicit guidelines for clinical characterization and diagnostic criteria.
Funding: This work was funded by core funds provided to the Viral Special Pathogens Branch at CDC.
Keywords: Bunyavirus; Disease surveillance; Genetics; HCPS; HFRS; HPS; Hantavirus cardiopulmonary syndrome; Hantavirus haemorrhagic fever with renal syndrome; Hantavirus pulmonary syndrome; Orthohantavirus; Rodent.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Viruses ICotTo. Bunyaviridae. https://ictv.global/report_9th/RNAneg/Bunyaviridae2023
-
- Lee H.W., French G.R., Lee P.W., Baek L.J., Tsuchiya K., Foulke R.S. Observations on natural and laboratory infection of rodents with the etiologic agent of Korean hemorrhagic fever. Am J Trop Med Hyg. 1981;30(2):477–482. - PubMed
-
- Enria D., Padula P., Segura E.L., et al. Hantavirus pulmonary syndrome in Argentina. Possibility of person to person transmission. Medicina (B Aires) 1996;56(6):709–711. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

