Analysis of Guard Cell Readouts Using Arabidopsis thaliana Isolated Epidermal Peels
- PMID: 39100596
- PMCID: PMC11292165
- DOI: 10.21769/BioProtoc.5033
Analysis of Guard Cell Readouts Using Arabidopsis thaliana Isolated Epidermal Peels
Abstract
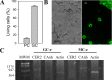
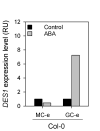
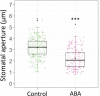
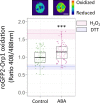
Stomata are pores surrounded by a pair of specialized cells, called guard cells, that play a central role in plant physiology through the regulation of gas exchange between plants and the environment. Guard cells have features like cell-autonomous responses and easily measurable readouts that have turned them into a model system to study signal transduction mechanisms in plants. Here, we provide a detailed protocol to analyze different physiological responses specifically in guard cells. We describe, in detail, the steps and conditions to isolate epidermal peels with tweezers and to analyze i) stomatal aperture in response to different stimuli, ii) cytosolic parameters such as hydrogen peroxide (H2O2), glutathione redox potential (E GSH), and MgATP-2 in vivo dynamics using fluorescent biosensors, and iii) gene expression in guard cell-enriched samples. The importance of this protocol lies in the fact that most living cells on epidermal peels are guard cells, enabling the preparation of guard cell-enriched samples. Key features • Isolation of epidermal peels as a monolayer enriched in guard cells • Measurement of cytosolic guard cell signaling component dynamics in isolated epidermal peels through fluorescent biosensor analysis • Gene expression analysis of guard cell-enriched isolated tissue.
Keywords: Arabidopsis thaliana; Biosensors; Epidermal peels; Guard cells; RNA; Stomatal aperture.
©Copyright : © 2024 The Authors; This is an open access article under the CC BY-NC license.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures

References
-
- WEYERS J. D. B. and TRAVIS A. J.(1981). Selection and Preparation of Leaf Epidermis for Experiments on Stomatal Physiology. J Exp Bot. 32(4): 837 850 850. 10.1093/jxb/32.4.837 - DOI
LinkOut - more resources
Full Text Sources

