Fine mapping of interspecific secondary CSSL populations revealed key regulators for grain weight at qTGW3.1 locus from Oryza nivara
- PMID: 39100880
- PMCID: PMC11291809
- DOI: 10.1007/s12298-024-01483-0
Fine mapping of interspecific secondary CSSL populations revealed key regulators for grain weight at qTGW3.1 locus from Oryza nivara
Abstract
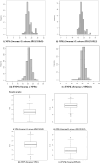
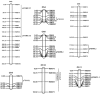
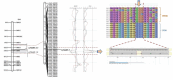
Grain weight (GW) is the most important stable trait that directly contributes to crop yield in case of cereals. A total of 105 backcross introgression lines (BC2F10 BILs) derived from Swarna/O. nivara IRGC81848 (NPS) and 90 BILs from Swarna/O. nivara IRGC81832 (NPK) were evaluated for thousand-grain weight (TGW) across four years (wet seasons 2014, 2015, 2016 and 2018) and chromosome segment substitution lines (CSSLs) were selected. From significant pair- wise mean comparison with Swarna, a total of 77 positively and 29 negatively significant NPS lines and 62 positively and 29 negatively significant NPK lines were identified. In all 4 years, 14 NPS lines and 9 NPK lines were positively significant and one-line NPS69 (IET22161) was negatively significant for TGW over Swarna consistently. NPS lines and NPK lines were genotyped using 111 and 140 polymorphic SSRs respectively. Quantitative trait locus (QTL) mapping using ICIM v4.2 software showed 13 QTLs for TGW in NPS. Three major effect QTLs qTGW2.1, qTGW8.1 and qTGW11.1 were identified in NPS for two or more years with PVE ranging from 8 to 14%. Likewise, 10 QTLs were identified in NPK and including two major effect QTL qTGW3.1 and qTGW12.1 with 6 to 32% PVE. In all QTLs, O. nivara alleles increased TGW. These consistent QTLs are very suitable for fine mapping and functional analysis of grain weight. Further in this study, CSSLs NPS1 (10-2S) and NPK61 (158 K) with significantly higher grain weight than the recurrent parent, Swarna cv. Oryza sativa were selected from each population and secondary F2 mapping populations were developed. Using Bulked Segregant QTL sequencing, a grain weight QTL, designated as qTGW3.1 was fine mapped from the cross between NPK61 and Swarna. This QTL explained 48% (logarithm of odds = 32.2) of the phenotypic variations and was fine mapped to a 31 kb interval using recombinant analysis. GRAS transcription factor gene (OS03go103400) involved in plant growth and development located at this genomic locus might be the candidate gene for qTGW3.1. The results of this study will help in further functional studies and improving the knowledge related to the molecular mechanism of grain weight in Oryza and lays a solid foundation for the breeding for high yield.
Supplementary information: The online version contains supplementary material available at 10.1007/s12298-024-01483-0.
Keywords: CSSLs; GBS; GRAS; Grain weight; Oryza nivara; QTLs.
© Prof. H.S. Srivastava Foundation for Science and Society 2024. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships and there is no potential conflict of interest.
Figures


Similar articles
-
Fine mapping of qROL1 for root length at early seedling stage from wild rice (Oryza nivara).Mol Breed. 2025 Apr 7;45(4):41. doi: 10.1007/s11032-025-01564-2. eCollection 2025 Apr. Mol Breed. 2025. PMID: 40206221
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Identification of Major Effect QTLs for Agronomic Traits and CSSLs in Rice from Swarna/Oryza nivara Derived Backcross Inbred Lines.Front Plant Sci. 2017 Jun 22;8:1027. doi: 10.3389/fpls.2017.01027. eCollection 2017. Front Plant Sci. 2017. PMID: 28690618 Free PMC article.
References
-
- Addanki KR, Balakrishnan D, Rao YV, Malathi S, Sukumar M, Kavitha B, Sarla N (2018) Swarna × Oryza nivara introgression lines: a resource for seedling vigour traits in rice. Plant Genet Resour UK 17(1):12–23 10.1017/S1479262118000187 - DOI
-
- Anilkumar C, Sah RP, Muhammed Azharudheen TP, Behera S, Singh N, Prakash NR, Sunitha NC, Devanna BN, Marndi BC, Patra BC, Nair SK (2022) Understanding complex genetic architecture of rice grain weight through QTL-meta analysis and candidate gene identification. Sci Rep 12(1):13832. 10.1038/s41598-022-17402-w 10.1038/s41598-022-17402-w - DOI - PMC - PubMed
-
- Balakrishnan D, Subrahmanyam D, Jyothi B, Raju AK, Rao YV, Kavitha B, Sukumar M, Malathi S, Revathi P, Padmavathi G, Ravindra Babu V, Sarla N (2016) Genotype × Environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara. Front Plant Sci 7:1530. 10.3389/fpls.2016.01530 10.3389/fpls.2016.01530 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
