Revealing the functional potential of microbial community of activated sludge for treating tuna processing wastewater through metagenomic analysis
- PMID: 39101040
- PMCID: PMC11294940
- DOI: 10.3389/fmicb.2024.1430199
Revealing the functional potential of microbial community of activated sludge for treating tuna processing wastewater through metagenomic analysis
Abstract
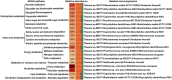
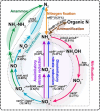
Reports regarding the composition and functions of microorganisms in activated sludge from wastewater treatment plants for treating tuna processing wastewater remains scarce, with prevailing studies focusing on municipal and industrial wastewater. This study delves into the efficiency and biological dynamics of activated sludge from tuna processing wastewater, particularly under conditions of high lipid content, for pollutant removal. Through metagenomic analysis, we dissected the structure of microbial community, and its relevant biological functions as well as pathways of nitrogen and lipid metabolism in activated sludge. The findings revealed the presence of 19 phyla, 1,880 genera, and 7,974 species, with Proteobacteria emerging as the predominant phylum. The study assessed the relative abundance of the core microorganisms involved in nitrogen removal, including Thauera sp. MZ1T and Alicycliphilus denitrificans K601, among others. Moreover, the results also suggested that a diverse array of fatty acid-degrading microbes, such as Thauera aminoaromatica and Cupriavidus necator H16, could thrive under lipid-rich conditions. This research can provide some referable information for insights into optimizing the operations of wastewater treatment and identify some potential microbial agents for nitrogen and fatty acid degradation.
Keywords: activated sludge; metagenomics; nitrogen removal; tuna processing; wastewater.
Copyright © 2024 Zheng, Liao, Chen, Ming, Jiao, Kong, Su and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Alves M. H., Campos-Takaki G. M., Porto A. L. F., Milanez A. I. (2002). Screening of Mucor spp. for the production of amylase, lipase, polygalacturonase and protease. Braz. J. Microbiol. 33, 325–330. doi: 10.1590/S1517-83822002000400009 - DOI
-
- Beldean-Galea M. S., Vial J., Thiébaut D., Coman V. (2013). Characterization of the fate of lipids in wastewater treatment using a comprehensive GCxGC/qMS and statistical approach. Anal. Methods 5, 2315–2323. doi: 10.1039/c3ay00060e - DOI
-
- Cao S. B., Cheng Z. Y., Koch K., Fang J. X., Du R., Peng Y. Z. (2024). Municipal wastewater driven partial-denitrification (PD) aggravated nitrous oxide (N2O) production. J. Clean. Prod. 434:139916. doi: 10.1016/j.jclepro.2023.139916 - DOI
LinkOut - more resources
Full Text Sources

