Revealing the role of metformin in gastric intestinal metaplasia treatment
- PMID: 39101145
- PMCID: PMC11294171
- DOI: 10.3389/fphar.2024.1340309
Revealing the role of metformin in gastric intestinal metaplasia treatment
Abstract
Objective: Gastric intestinal metaplasia (IM) is a precancerous stage associated with gastric cancer. Despite the observed beneficial effects of metformin on IM, its molecular mechanism remains not fully elucidated. This study aims to reveal the effects and potential mechanisms of metformin in treating IM based on both bioinformatics and in vivo investigations.
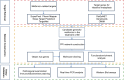
Methods: The seven public databases (GeneCards, DisGeNET, OMIM, SuperPred, Pharm Mapper, Swiss Target Prediction, TargetNet) were used in this work to identify targeted genes related to intestinal metaplasia (IM) and metformin. The shared targeted genes between metformin and IM were further analyzed by network pharmacology, while the interactions in-between were investigated by molecular docking. In parallel, the therapeutic effect of metformin was evaluated in IM mice model, while the core targets and pathways effected by metformin were verified in vivo.
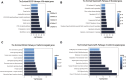
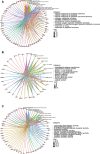
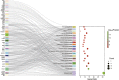
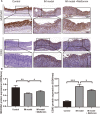
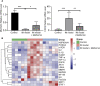
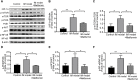
Results: We screened out 1,751 IM-related genes and 318 metformin-targeted genes, 99 common genes identified in between were visualized by constructing the protein-protein interaction (PPI) network. The top ten core targeted genes were EGFR, MMP9, HIF1A, HSP90AA1, SIRT1, IL2, MAPK8, STAT1, PIK3CA, and ICAM1. The functional enrichment analysis confirmed that carcinogenesis and HIF-1 signaling pathways were primarily involved in the metformin treatment of IM. Based on molecular docking and dynamics, we found metformin affected the function of its targets by inhibiting receptor binding. Furthermore, metformin administration reduced the progression of IM lesions in Atp4a-/- mice model significantly. Notably, metformin enhanced the expression level of MUC5AC, while inhibited the expression level of CDX2. Our results also showed that metformin modulated the expression of core targets in vivo by reducing the activity of NF-κB and the PI3K/AKT/mTOR/HIF-1α signaling pathway.
Conclusion: This study confirms that metformin improves the efficacy of IM treatment by regulating a complex molecular network. Metformin plays a functional role in inhibiting inflammation/apoptosis-related pathways of further IM progression. Our work provides a molecular foundation for understanding metformin and other guanidine medicines in IM treatment.
Keywords: intestinal metaplasia; metformin; molecular docking; network pharmacology; precancerous diseases.
Copyright © 2024 Hu, Xue, Sun, Mi, Wen, Xi, Li, Zheng and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Exploring the therapeutic mechanisms of Coptidis Rhizoma in gastric precancerous lesions: a network pharmacology approach.Discov Oncol. 2024 Jun 5;15(1):211. doi: 10.1007/s12672-024-01070-5. Discov Oncol. 2024. PMID: 38837097 Free PMC article.
-
Exploring the therapeutic mechanism of curcumin in prostate cancer using network pharmacology and molecular docking.Heliyon. 2024 Jun 19;10(12):e33103. doi: 10.1016/j.heliyon.2024.e33103. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 39022084 Free PMC article.
-
Mechanism of Bazhen decoction in the treatment of colorectal cancer based on network pharmacology, molecular docking, and experimental validation.Front Immunol. 2023 Sep 20;14:1235575. doi: 10.3389/fimmu.2023.1235575. eCollection 2023. Front Immunol. 2023. PMID: 37799727 Free PMC article.
-
Exploring the mechanism of metformin action in Alzheimer's disease and type 2 diabetes based on network pharmacology, molecular docking, and molecular dynamic simulation.Ther Adv Endocrinol Metab. 2023 Sep 27;14:20420188231187493. doi: 10.1177/20420188231187493. eCollection 2023. Ther Adv Endocrinol Metab. 2023. PMID: 37780174 Free PMC article.
-
Exploring the mechanism of curcumin in the treatment of colon cancer based on network pharmacology and molecular docking.Front Pharmacol. 2023 Feb 15;14:1102581. doi: 10.3389/fphar.2023.1102581. eCollection 2023. Front Pharmacol. 2023. PMID: 36874006 Free PMC article.
Cited by
-
Genetically engineered mouse models in gastric precancerous lesions research.World J Gastrointest Surg. 2025 Jul 27;17(7):107610. doi: 10.4240/wjgs.v17.i7.107610. World J Gastrointest Surg. 2025. PMID: 40740928 Free PMC article. Review.
References
-
- Ala M., Ala M. (2021). Metformin for cardiovascular protection, inflammatory bowel disease, osteoporosis, periodontitis, polycystic ovarian syndrome, neurodegeneration, cancer, inflammation and senescence: what is next? ACS Pharmacol. Transl. Sci. 4, 1747–1770. 10.1021/acsptsci.1c00167 - DOI - PMC - PubMed
-
- Cheng J., Li C., Ying Y., Lv J., Qu X., Mcgowan E., et al. (2022). Metformin alleviates endometriosis and potentiates endometrial receptivity via decreasing VEGF and MMP9 and increasing leukemia inhibitor factor and HOXA10. Front. Pharmacol. 13, 750208. 10.3389/fphar.2022.750208 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

