The immune landscape of the inflamed joint defined by spectral flow cytometry
- PMID: 39101538
- PMCID: PMC11557149
- DOI: 10.1093/cei/uxae071
The immune landscape of the inflamed joint defined by spectral flow cytometry
Abstract
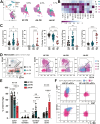
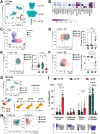
Cellular phenotype and function are altered in different microenvironments. For targeted therapies it is important to understand site-specific cellular adaptations. Juvenile idiopathic arthritis (JIA) is characterized by autoimmune joint inflammation, with frequent inadequate treatment responses. To comprehensively assess the inflammatory immune landscape, we designed a 37-parameter spectral flow cytometry panel delineating mononuclear cells from JIA synovial fluid (SF) of autoimmune inflamed joints, compared to JIA and healthy control blood. Synovial monocytes and NK cells (CD56bright) lack Fc-receptor CD16, suggesting antibody-mediated targeting may be ineffective. B cells and DCs, both in small frequencies in SF, undergo maturation with high 4-1BB, CD71, CD39 expression, supporting T-cell activation. SF effector and regulatory T cells were highly active with newly described co-receptor combinations that may alter function, and suggestion of metabolic reprogramming via CD71, TNFR2, and PD-1. Most SF effector phenotypes, as well as an identified CD4-Foxp3+ T-cell population, were restricted to the inflamed joint, yet specific SF-predominant CD4+ Foxp3+ Treg subpopulations were increased in blood of active but not inactive JIA, suggesting possible recirculation and loss of immunoregulation at distal sites. This first comprehensive dataset of the site-specific inflammatory landscape at protein level will inform functional studies and the development of targeted therapeutics to restore immunoregulatory balance and achieve remission in JIA.
Keywords: autoimmunity; cellular adaptations; inflamed joint; juvenile idiopathic arthritis; spectral flow cytometry.
© The Author(s) 2024. Published by Oxford University Press on behalf of the British Society for Immunology.
Conflict of interest statement
M.H.A., D.S., and A.M.P. have no conflict of interest to declare. L.R.W., M.K. and V.A. received funding/funding in kind to CLUSTER-JIA Consortium from Pfizer, UCB, AbbVie, SOBI and GSK, but that support did not directly fund this work. L.R.W. has received speaker fees, paid to UCL, from Pfizer, unrelated to this work.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

