Variation in the Spectrum of New Mutations among Inbred Strains of Mice
- PMID: 39101589
- PMCID: PMC11327921
- DOI: 10.1093/molbev/msae163
Variation in the Spectrum of New Mutations among Inbred Strains of Mice
Abstract
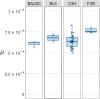
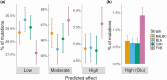
The mouse serves as a mammalian model for understanding the nature of variation from new mutations, a question that has both evolutionary and medical significance. Previous studies suggest that the rate of single-nucleotide mutations (SNMs) in mice is ∼50% of that in humans. However, information largely comes from studies involving the C57BL/6 strain, and there is little information from other mouse strains. Here, we study the mutations that accumulated in 59 mouse lines derived from four inbred strains that are commonly used in genetics and clinical research (BALB/cAnNRj, C57BL/6JRj, C3H/HeNRj, and FVB/NRj), maintained for eight to nine generations by brother-sister mating. By analyzing Illumina whole-genome sequencing data, we estimate that the average rate of new SNMs in mice is ∼μ = 6.7 × 10-9. However, there is substantial variation in the spectrum of SNMs among strains, so the burden from new mutations also varies among strains. For example, the FVB strain has a spectrum that is markedly skewed toward C→A transversions and is likely to experience a higher deleterious load than other strains, due to an increased frequency of nonsense mutations in glutamic acid codons. Finally, we observe substantial variation in the rate of new SNMs among DNA sequence contexts, CpG sites, and their adjacent nucleotides playing an important role.
Keywords: Mus musculus; mutation accumulation; mutation rate; mutation spectrum.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Alexaki A, Kames J, Holcomb DD, Athey J, Santana-Quintero LV, Lam PVN, Hamasaki-Katagiri N, Osipova E, Simonyan V, Bar H, et al. . Codon and codon-pair usage tables (CoCoPUTs): facilitating genetic variation analyses and recombinant gene design. J Mol Biol. 2019:431(13):2434–2441. 10.1016/j.jmb.2019.04.021. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

