Dynamic development of microglia and macrophages after spinal cord injury
- PMID: 39101644
- PMCID: PMC11974661
- DOI: 10.4103/NRR.NRR-D-24-00063
Dynamic development of microglia and macrophages after spinal cord injury
Abstract
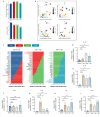
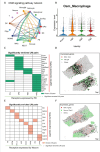
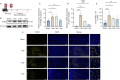
JOURNAL/nrgr/04.03/01300535-202512000-00029/figure1/v/2025-01-31T122243Z/r/image-tiff Secondary injury following spinal cord injury is primarily characterized by a complex inflammatory response, with resident microglia and infiltrating macrophages playing pivotal roles. While previous studies have grouped these two cell types together based on similarities in structure and function, an increasing number of studies have demonstrated that microglia and macrophages exhibit differences in structure and function and have different effects on disease processes. In this study, we used single-cell RNA sequencing and spatial transcriptomics to identify the distinct evolutionary paths of microglia and macrophages following spinal cord injury. Our results showed that microglia were activated to a pro-inflammatory phenotype immediately after spinal cord injury, gradually transforming to an anti-inflammatory steady state phenotype as the disease progressed. Regarding macrophages, our findings highlighted abundant communication with other cells, including fibroblasts and neurons. Both pro-inflammatory and neuroprotective effects of macrophages were also identified; the pro-inflammatory effect may be related to integrin β2 ( Itgb2 ) and the neuroprotective effect may be related to the oncostatin M pathway. These findings were validated by in vivo experiments. This research underscores differences in the cellular dynamics of microglia and macrophages following spinal cord injury, and may offer new perspectives on inflammatory mechanisms and potential therapeutic targets.
Copyright © 2025 Neural Regeneration Research.
Conflict of interest statement
Figures




References
-
- Anderson MA, O’Shea TM, Burda JE, Ao Y, Barlatey SL, Bernstein AM, Kim JH, James ND, Rogers A, Kato B, Wollenberg AL, Kawaguchi R, Coppola G, Wang C, Deming TJ, He Z, Courtine G, Sofroniew MV. Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature. 2018;561:396–400. - PMC - PubMed
-
- Badhiwala JH, Wilson JR, Fehlings MG. Global burden of traumatic brain and spinal cord injury. Lancet Neurol. 2019;18:24–25. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

