Isolation and characterization of Septuagintavirus; a novel clade of Escherichia coli phages within the subfamily Vequintavirinae
- PMID: 39101714
- PMCID: PMC11370258
- DOI: 10.1128/spectrum.00592-24
Isolation and characterization of Septuagintavirus; a novel clade of Escherichia coli phages within the subfamily Vequintavirinae
Abstract
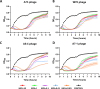
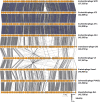
Escherichia coli is a commensal inhabitant of the mammalian gut microbiota, frequently associated with various gastrointestinal diseases. There is increasing interest in comprehending the variety of bacteriophages (phages) that target this bacterium, as such insights could pave the way for their potential use in therapeutic applications. Here, we report the isolation and characterization of four newly identified E. coli infecting tailed phages (W70, A7-1, A5-4, and A73) that were found to constitute a novel genus, Septuagintavirus, within the subfamily Vequintavirinae. Genomes of these phages ranged from 137 kbp to 145 kbp, with a GC content of 41 mol%. They possess a maximum nucleotide similarity of 30% with phages of the closest phylogenetic genus, Certrevirus, while displaying limited homology to other genera of the Vequintavirinae family. Host range analysis showed that these phages have limited activity against a panel of E. coli strains, infecting 6 out of 16 tested isolates, regardless of their phylotype. Electrospray ionization-tandem mass spectrometry (ESI-MS/MS) was performed on the virion of phage W70, allowing the identification of 28 structural proteins, 19 of which were shared with phages of other genera of Vequintavirinae family. The greatest diversity was identified with proteins forming tail fiber structures, likely indicating the adaptation of virions of each phage genus of this subfamily for the recognition of their target receptor on host cells. The findings of this study provide greater insights into the phages of the subfamily Vequintavirinae, contributing to the pool of knowledge currently known about these phages.
Importance: Escherichia coli is a well-known bacterium that inhabits diverse ecological niches, including the mammalian gut microbiota. Certain strains are associated with gastrointestinal diseases, and there is a growing interest in using bacteriophages, viruses that infect bacteria, to combat bacterial infections. Here, we describe the isolation and characterization of four novel E. coli bacteriophages that constitute a new genus, Septuagintavirus, within the subfamily Vequintavirinae. We conducted mass spectrometry on virions of a representative phage of this novel clade and compared it to other phages within the subfamily. Our analysis shows that virion structure is highly conserved among all phages, except for proteins related to tail fiber structures implicated in the host range. These findings provide greater insights into the phages of the subfamily Vequintavirinae, contributing to the existing pool of knowledge about these phages.
Keywords: Escherichia coli; Vequintavirinae; bacteriophage; characterization; isolation; mass-spectrometry; virion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Isolation and Characterization of Pectobacterium Phage vB_PatM_CB7: New Insights into the Genus Certrevirus.Antibiotics (Basel). 2020 Jun 21;9(6):352. doi: 10.3390/antibiotics9060352. Antibiotics (Basel). 2020. PMID: 32575906 Free PMC article.
-
The Isolation and Characterization of Bacteriophages Infecting Avian Pathogenic Escherichia coli O1, O2 and O78 Strains.Viruses. 2023 Oct 16;15(10):2095. doi: 10.3390/v15102095. Viruses. 2023. PMID: 37896873 Free PMC article.
-
A Quest of Great Importance-Developing a Broad Spectrum Escherichia coli Phage Collection.Viruses. 2019 Sep 26;11(10):899. doi: 10.3390/v11100899. Viruses. 2019. PMID: 31561510 Free PMC article.
-
Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key "Blueprint" for Reprogramming Phage Host Range.Int J Mol Sci. 2022 Oct 12;23(20):12146. doi: 10.3390/ijms232012146. Int J Mol Sci. 2022. PMID: 36292999 Free PMC article. Review.
-
Structure, Biology, and Applications of Filamentous Bacteriophages.Cold Spring Harb Protoc. 2024 Aug 1;2024(8):pdb.over107754. doi: 10.1101/pdb.over107754. Cold Spring Harb Protoc. 2024. PMID: 37460152 Review.
Cited by
-
Physicochemical, genomic, and phenotypic characterization of Escherichia phage BME3.Microbiol Spectr. 2025 Jul;13(7):e0130124. doi: 10.1128/spectrum.01301-24. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401929 Free PMC article.
References
-
- Nadalian B, Yadegar A, Houri H, Olfatifar M, Shahrokh S, Asadzadeh Aghdaei H, Suzuki H, Zali MR. 2021. Prevalence of the pathobiont adherent-invasive Escherichia coli and inflammatory bowel disease: a systematic review and meta-analysis. J Gastroenterol Hepatol 36:852–863. doi: 10.1111/jgh.15260 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

