Bifidobacterium and Lactobacillus Probiotics and Gut Dysbiosis in Preterm Infants: The PRIMAL Randomized Clinical Trial
- PMID: 39102225
- PMCID: PMC12549143
- DOI: 10.1001/jamapediatrics.2024.2626
Bifidobacterium and Lactobacillus Probiotics and Gut Dysbiosis in Preterm Infants: The PRIMAL Randomized Clinical Trial
Abstract
Importance: The effects of probiotic interventions on colonization with resistant bacteria and early microbiome development in preterm infants remain to be clarified.
Objective: To examine the efficacy of Bifidobacterium longum subsp infantis, Bifidobacterium animalis subsp lactis (BB-12), and Lactobacillus acidophilus (La-5) probiotics to prevent colonization with multidrug-resistant organisms or highly epidemic bacteria (MDRO+) and to shape the microbiome of preterm infants toward the eubiotic state of healthy full-term infants.
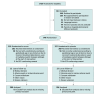
Design, setting, and participants: The multicenter, double-blinded, placebo-controlled, group sequential, phase 3 Priming Immunity at the Beginning of Life (PRIMAL) randomized clinical trial, conducted from April 2018 to June 2020, included infants with gestational age of 28 to 32 weeks at 18 German neonatal units. Data analyses were conducted from March 2020 to August 2023.
Intervention: A total of 28 days of multistrain probiotics diluted in human milk/formula starting within the first 72 hours of life.
Main outcomes and measures: Colonization with MDRO+ at day 30 of life (primary end point), late-onset sepsis and severe gastrointestinal complication (safety end points), and gut dysbiosis, ie, deviations from the microbiome of healthy, term infants (eubiosis score) based on 16-subunit ribosomal RNA and metagenomic sequencing.
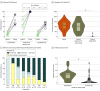
Results: Among the 643 infants randomized until the stop of recruitment based on interim results, 618 (median [IQR] gestational age, 31.0 [29.7-32.1] weeks; 333 male [53.9%]; mean [SD] birth weight, 1502 [369] g) had follow-up at day 30. The interim analysis with all available data from 219 infants revealed MDRO+ colonization in 43 of 115 infants (37.4%) in the probiotics group and in 39 of 104 infants (37.5%) in the control group (adjusted risk ratio, 0.99; 95% CI, 0.54-1.81; P = .97). Safety outcomes were similar in both groups, ie, late-onset sepsis (probiotics group: 8 of 316 infants [2.5%]; control group: 12 of 322 infants [3.7%]) and severe gastrointestinal complications (probiotics group: 6 of 316 infants [1.9%]; control group: 7 of 322 infants [2.2%]). The probiotics group had higher eubiosis scores than the control group at the genus level (254 vs 258 infants; median scores, 0.47 vs 0.41; odds ratio [OR], 1.07; 95% CI, 1.02-1.13) and species level (96 vs 83 infants; median scores, 0.87 vs 0.59; OR, 1.28; 95% CI, 1.19-1.38). Environmental uptake of the B infantis probiotic strain in the control group was common (41 of 84 [49%]), which was highly variable across sites and particularly occurred in infants with a sibling who was treated with probiotics.
Conclusions and relevance: Multistrain probiotics did not reduce the incidence of MDRO+ colonization at day 30 of life in preterm infants but modulated their microbiome toward eubiosis.
Trial registration: German Clinical Trials Register: DRKS00013197.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

