Genome-wide association study of the loci and candidate genes associated with agronomic traits in Amomum villosum Lour
- PMID: 39102408
- PMCID: PMC11299815
- DOI: 10.1371/journal.pone.0306806
Genome-wide association study of the loci and candidate genes associated with agronomic traits in Amomum villosum Lour
Abstract
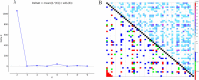
Amomum villosum Lour. (A. villosum) is a valuable herbaceous plant that produces the famous traditional Chinese medicine Amori Fructus. Identifying molecular markers associated with the growth of A. villosum can facilitate molecular marker-assisted breeding of the plant. This study employed 75 A. villosum accessions as the test material and utilized 71 pairs of polymorphic simple sequence repeat (SSR) molecular markers to genotype the population. The study analyzed the association between SSR markers and phenotypic traits through the linkage imbalance and population structure analysis. Candidate genes associated with the molecular markers were also identified. The results showed that the phenotypic diversity index range of the 12 agronomic traits was 4.081-4.312 and conformed to a normal distribution. Moreover, 293 allelic variations were detected in the 75 accessions, with an average of 5.32 amplified alleles per loci, ranging from 3 to 8. The maximum number of amplified alleles for AVL12 was 8. The population structure and cluster analysis indicated that the accessions could be divided into two subgroups. Using the mixed linear model (MLM) model of population structure (Q)+kinship matrix (K) for association analysis, three SSR molecular markers significantly associated with the agronomic traits were detected. Fluorescence quantification was used to analyze the expression levels of six candidate genes, and it was found that three of the genes were differentially expressed in phenotypically different accessions. This study is the first to use SSR markers for genome-wide association study (GWAS) mapping and identification of the associated agronomic traits in A. villosum. The results of this study provide a basis for identifying genetic markers for growth traits for marker-assisted breeding in A. villosum.
Copyright: © 2024 Li et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Xuefeng X, Xueying LI, Feilong C, Wenxue XU. Study on Chemical Constituents of Essential Oil from Fruits and Leaves of Amomum villosum Lour. by GC-MS. Traditional Chinese Drug Research and Clinical Pharmacology. 2012;5(5): 760–765. doi: 10.4236/ajps.2014.55090 - DOI
-
- De Boer H, Newman M, Poulsen AD, Droop AJ, Fér T, Hiên LTT, et al. Convergent morphology in Alpinieae (Zingiberaceae): Recircumscribing Amomum as a monophyletic genus. Taxon. 2018;67(1): 6–36. doi: 10.12705/671.2 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

