Comparative analysis of new mScarlet-based red fluorescent tags in Caenorhabditis elegans
- PMID: 39103170
- PMCID: PMC11457934
- DOI: 10.1093/genetics/iyae126
Comparative analysis of new mScarlet-based red fluorescent tags in Caenorhabditis elegans
Abstract
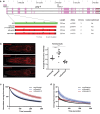
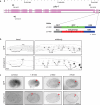
One problem that has hampered the use of red fluorescent proteins in the fast-developing nematode Caenorhabditis elegans has been the substantial time delay in maturation of several generations of red fluorophores. The recently described mScarlet-I3 protein has properties that may overcome this limitation. We compare here the brightness and onset of expression of CRISPR/Cas9 genome-engineered mScarlet, mScarlet3, mScarlet-I3, and GFP reporter knock-ins. Comparing the onset and brightness of expression of reporter alleles of C. elegans golg-4, encoding a broadly expressed Golgi resident protein, we found that the onset of detection of mScarlet-I3 in the embryo is several hours earlier than older versions of mScarlet and comparable to GFP. These findings were further supported by comparing mScarlet-I3 and GFP reporter alleles for pks-1, a gene expressed in the CAN neuron and cells of the alimentary system, as well as reporter alleles for the pan-neuronal, nuclear marker unc-75. Hence, the relative properties of mScarlet-I3 and GFP do not depend on cellular or subcellular context. In all cases, mScarlet-I3 reporters also show improved signal-to-noise ratio compared to GFP.
Keywords: Caenorhabditis elegans; RFP; fluorophore; marker.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures


Update of
-
Comparative analysis of new, mScarlet-based red fluorescent tags in Caenorhabditis elegans.bioRxiv [Preprint]. 2024 Jun 13:2024.06.11.598534. doi: 10.1101/2024.06.11.598534. bioRxiv. 2024. Update in: Genetics. 2024 Oct 7;228(2):iyae126. doi: 10.1093/genetics/iyae126. PMID: 38915494 Free PMC article. Updated. Preprint.
References
-
- El Mouridi S, Lecroisey C, Tardy P, Mercier M, Leclercq-Blondel A, Zariohi N, Boulin T. 2017. Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype. G3 (Bethesda). 7(5):1429–1437. doi: 10.1534/g3.117.040824. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

