Functional genetics reveals the contribution of delta opioid receptor to type 2 diabetes and beta-cell function
- PMID: 39103322
- PMCID: PMC11300616
- DOI: 10.1038/s41467-024-51004-6
Functional genetics reveals the contribution of delta opioid receptor to type 2 diabetes and beta-cell function
Abstract
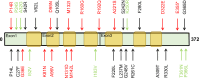
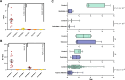
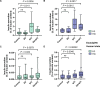
Functional genetics has identified drug targets for metabolic disorders. Opioid use impacts metabolic homeostasis, although mechanisms remain elusive. Here, we explore the OPRD1 gene (encoding delta opioid receptor, DOP) to understand its impact on type 2 diabetes. Large-scale sequencing of OPRD1 and in vitro analysis reveal that loss-of-function variants are associated with higher adiposity and lower hyperglycemia risk, whereas gain-of-function variants are associated with lower adiposity and higher type 2 diabetes risk. These findings align with studies of opium addicts. OPRD1 is expressed in human islets and beta cells, with decreased expression under type 2 diabetes conditions. DOP inhibition by an antagonist enhances insulin secretion from human beta cells and islets. RNA-sequencing identifies pathways regulated by DOP antagonism, including nerve growth factor, circadian clock, and nuclear receptor pathways. Our study highlights DOP as a key player between opioids and metabolic homeostasis, suggesting its potential as a therapeutic target for type 2 diabetes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

