Distinct horizontal transfer mechanisms for type I and type V CRISPR-associated transposons
- PMID: 39103341
- PMCID: PMC11300857
- DOI: 10.1038/s41467-024-50816-w
Distinct horizontal transfer mechanisms for type I and type V CRISPR-associated transposons
Abstract
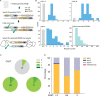
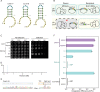
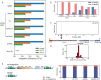
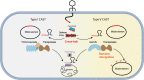
CASTs use both CRISPR-associated proteins and Tn7-family transposons for RNA-guided vertical and horizontal transmission. CASTs encode minimal CRISPR arrays but can't acquire new spacers. Here, we report that CASTs can co-opt defense-associated CRISPR arrays for horizontal transmission. A bioinformatic analysis shows that CASTs co-occur with defense-associated CRISPR systems, with the highest prevalence for type I-B and type V CAST sub-types. Using an E. coli quantitative transposition assay and in vitro reconstitution, we show that CASTs can use CRISPR RNAs from these defense systems. A high-resolution structure of the type I-F CAST-Cascade in complex with a type III-B CRISPR RNA reveals that Cas6 recognizes direct repeats via sequence-independent π - π interactions. In addition to using heterologous CRISPR arrays, type V CASTs can also transpose via an unguided mechanism, even when the S15 co-factor is over-expressed. Over-expressing S15 and the trans-activating CRISPR RNA or a single guide RNA reduces, but does not abrogate, off-target integration for type V CASTs. Our findings suggest that some CASTs may exploit defense-associated CRISPR arrays and that this fact must be considered when porting CASTs to heterologous bacterial hosts. More broadly, this work will guide further efforts to engineer the activity and specificity of CASTs for gene editing applications.
© 2024. The Author(s).
Conflict of interest statement
K.H., C.O.W., I.J.F., and UT-Austin have filed a patent disclosure relating to using CRISPR-associated transposons for bacterial and mammalian gene editing. The remaining authors declare no competing interests.
Figures


Update of
-
Distinct horizontal transfer mechanisms for type I and type V CRISPR-associated transposons.bioRxiv [Preprint]. 2023 Jul 11:2023.03.03.531003. doi: 10.1101/2023.03.03.531003. bioRxiv. 2023. Update in: Nat Commun. 2024 Aug 6;15(1):6653. doi: 10.1038/s41467-024-50816-w. PMID: 37502928 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 GM124141/GM/NIGMS NIH HHS/United States
- R01GM088344/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 GM088344/GM/NIGMS NIH HHS/United States
- F-1808/Welch Foundation
- R01GM124141/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

