Plasmodium falciparum population dynamics in East Africa and genomic surveillance along the Kenya-Uganda border
- PMID: 39103358
- PMCID: PMC11300580
- DOI: 10.1038/s41598-024-67623-4
Plasmodium falciparum population dynamics in East Africa and genomic surveillance along the Kenya-Uganda border
Abstract
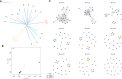
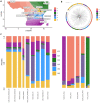
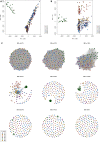
East African countries accounted for ~ 10% of all malaria prevalence worldwide in 2022, with an estimated 23.8 million cases and > 53,000 deaths. Despite recent increases in malaria incidence, high-resolution genome-wide analyses of Plasmodium parasite populations are sparse in Kenya, Tanzania, and Uganda. The Kenyan-Ugandan border region is a particular concern, with Uganda confirming the emergence and spread of artemisinin resistant P. falciparum parasites. To establish genomic surveillance along the Kenyan-Ugandan border and analyse P. falciparum population dynamics within East Africa, we generated whole-genome sequencing (WGS) data for 38 parasites from Bungoma, Western Kenya. These sequences were integrated into a genomic analysis of available East African isolate data (n = 599) and revealed parasite subpopulations with distinct genetic structure and diverse ancestral origins. Ancestral admixture analysis of these subpopulations alongside isolates from across Africa (n = 365) suggested potential independent ancestral populations from other major African populations. Within isolates from Western Kenya, the prevalence of biomarkers associated with chloroquine resistance (e.g. Pfcrt K76T) were significantly reduced compared to wider East African populations and a single isolate contained the PfK13 V568I variant, potentially linked to reduced susceptibility to artemisinin. Overall, our work provides baseline WGS data and analysis for future malaria genomic surveillance in the region.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Geneva: World Health Organization (2022) World malaria report 2022. Licence: CC BY-NC-SA 3.0 IGO
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

