A comprehensive single-cell RNA transcriptomic analysis identifies a unique SPP1+ macrophages subgroup in aging skeletal muscle
- PMID: 39103421
- PMCID: PMC11300837
- DOI: 10.1038/s41598-024-69284-9
A comprehensive single-cell RNA transcriptomic analysis identifies a unique SPP1+ macrophages subgroup in aging skeletal muscle
Abstract
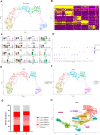
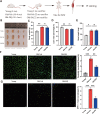
Senescence of skeletal muscle (SkM) has been a primary contributor to senior weakness and disability in recent years. The gradually declining SkM function associated with senescence has recently been connected to an imbalance between damage and repair. Macrophages (Mac) are involved in SkM aging, and different macrophage subgroups hold different biological functions. Through comprehensive single-cell transcriptomic analysis, we first compared the metabolic pathways and biological functions of different types of cells in young (Y) and old (O) mice SkM. Strikingly, the Mac population in mice SkM was also explored, and we identified a unique Mac subgroup in O SkM characterized by highly expressed SPP1 with strong senescence and adipogenesis features. Further work was carried out on the metabolic and biological processes for these Mac subgroups. Besides, we verified that the proportion of the SPP1+ Mac was increased significantly in the quadriceps tissues of O mice, and the senotherapeutic drug combination dasatinib + quercetin (D + Q) could dramatically reduce its proportion. Our study provides novel insight into the potential role of SPP1+ Mac in SkM, which may serve as a senotherapeutic target in SkM aging.
Keywords: Cell senescence; SPP1+ macrophages; Senolytics; Single-cell sequencing; Skeletal muscle.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

