Rapid, biochemical tagging of cellular activity history in vivo
- PMID: 39103446
- PMCID: PMC11399108
- DOI: 10.1038/s41592-024-02375-7
Rapid, biochemical tagging of cellular activity history in vivo
Abstract
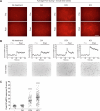
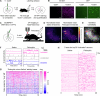
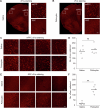
Intracellular calcium (Ca2+) is ubiquitous to cell signaling across biology. While existing fluorescent sensors and reporters can detect activated cells with elevated Ca2+ levels, these approaches require implants to deliver light to deep tissue, precluding their noninvasive use in freely behaving animals. Here we engineered an enzyme-catalyzed approach that rapidly and biochemically tags cells with elevated Ca2+ in vivo. Ca2+-activated split-TurboID (CaST) labels activated cells within 10 min with an exogenously delivered biotin molecule. The enzymatic signal increases with Ca2+ concentration and biotin labeling time, demonstrating that CaST is a time-gated integrator of total Ca2+ activity. Furthermore, the CaST readout can be performed immediately after activity labeling, in contrast to transcriptional reporters that require hours to produce signal. These capabilities allowed us to apply CaST to tag prefrontal cortex neurons activated by psilocybin, and to correlate the CaST signal with psilocybin-induced head-twitch responses in untethered mice.
© 2024. The Author(s).
Conflict of interest statement
D.E.O. is a cofounder of Delix Therapeutics and serves as the chief innovation officer and head of the scientific advisory board. The other authors declare no competing interests.
Figures




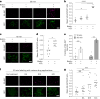

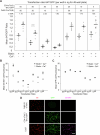




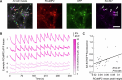
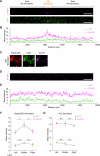
Update of
-
Rapid, biochemical tagging of cellular activity history in vivo.bioRxiv [Preprint]. 2024 May 14:2023.09.06.556431. doi: 10.1101/2023.09.06.556431. bioRxiv. 2024. Update in: Nat Methods. 2024 Sep;21(9):1725-1735. doi: 10.1038/s41592-024-02375-7. PMID: 38798353 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- NRT NeuralStorm #2152260/National Science Foundation (NSF)
- NARSAD Young Investigator Grant #30238/Brain and Behavior Research Foundation (Brain & Behavior Research Foundation)
- DP2 MH136588/MH/NIMH NIH HHS/United States
- R35 GM148182/GM/NIGMS NIH HHS/United States
- T32 MH112507/MH/NIMH NIH HHS/United States
- R35GM148182/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- Searle Scholars Program #SSP-2022-107/Kinship Foundation
- T32 MH082174/MH/NIMH NIH HHS/United States
- DP2MH136588/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- Beckman Young Investigator Award/Arnold and Mabel Beckman Foundation
- GRFP #000895154/National Science Foundation (NSF)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

