Real-time assessment of mitochondrial DNA heteroplasmy dynamics at the single-cell level
- PMID: 39103491
- PMCID: PMC11574196
- DOI: 10.1038/s44318-024-00183-5
Real-time assessment of mitochondrial DNA heteroplasmy dynamics at the single-cell level
Abstract
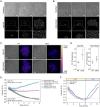
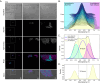
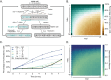
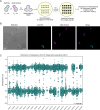
Mitochondrial DNA (mtDNA) is present in multiple copies within cells and is required for mitochondrial ATP generation. Even within individual cells, mtDNA copies can differ in their sequence, a state known as heteroplasmy. The principles underlying dynamic changes in the degree of heteroplasmy remain incompletely understood, due to the inability to monitor this phenomenon in real time. Here, we employ mtDNA-based fluorescent markers, microfluidics, and automated cell tracking, to follow mtDNA variants in live heteroplasmic yeast populations at the single-cell level. This approach, in combination with direct mtDNA tracking and data-driven mathematical modeling reveals asymmetric partitioning of mtDNA copies during cell division, as well as limited mitochondrial fusion and fission frequencies, as critical driving forces for mtDNA variant segregation. Given that our approach also facilitates assessment of segregation between intact and mutant mtDNA, we anticipate that it will be instrumental in elucidating the mechanisms underlying the purifying selection of mtDNA.
Keywords: Heteroplasmy; Mathematical Modeling; Mitochondria; Mitochondrial Fission; mtDNA.
© 2024. The Author(s).
Conflict of interest statement
Figures



Similar articles
-
Single-cell analysis reveals context-dependent, cell-level selection of mtDNA.Nature. 2024 May;629(8011):458-466. doi: 10.1038/s41586-024-07332-0. Epub 2024 Apr 24. Nature. 2024. PMID: 38658765 Free PMC article.
-
Tissue-specific modulation of mitochondrial DNA segregation by a defect in mitochondrial division.Hum Mol Genet. 2016 Feb 15;25(4):706-14. doi: 10.1093/hmg/ddv508. Epub 2015 Dec 17. Hum Mol Genet. 2016. PMID: 26681804
-
Mutant C. elegans mitofusin leads to selective removal of mtDNA heteroplasmic deletions across generations to maintain fitness.BMC Biol. 2022 Feb 9;20(1):40. doi: 10.1186/s12915-022-01241-2. BMC Biol. 2022. PMID: 35139855 Free PMC article.
-
Cellular mechanisms of mtDNA heteroplasmy dynamics.Crit Rev Biochem Mol Biol. 2021 Oct;56(5):510-525. doi: 10.1080/10409238.2021.1934812. Epub 2021 Jun 13. Crit Rev Biochem Mol Biol. 2021. PMID: 34120542 Review.
-
Rolling-Circle Replication in Mitochondrial DNA Inheritance: Scientific Evidence and Significance from Yeast to Human Cells.Genes (Basel). 2020 May 6;11(5):514. doi: 10.3390/genes11050514. Genes (Basel). 2020. PMID: 32384722 Free PMC article. Review.
Cited by
-
The Volume of Mitochondria Inherited Impacts mtDNA Homeostasis in Budding Yeast.bioRxiv [Preprint]. 2025 Mar 26:2025.03.25.645216. doi: 10.1101/2025.03.25.645216. bioRxiv. 2025. PMID: 40196573 Free PMC article. Preprint.
-
The power of connections: Recent advances in understanding the regulation of mitochondrial dynamics by membrane contact sites.Curr Opin Cell Biol. 2025 Aug;95:102535. doi: 10.1016/j.ceb.2025.102535. Epub 2025 May 28. Curr Opin Cell Biol. 2025. PMID: 40441071 Review.
-
Inheritance bias of deletion-harbouring mtDNA in yeast: The role of copy number and intracellular selection.PLoS Genet. 2025 Jun 24;21(6):e1011737. doi: 10.1371/journal.pgen.1011737. eCollection 2025 Jun. PLoS Genet. 2025. PMID: 40554585 Free PMC article.
-
Mitochondrial curation for the next generation.Curr Opin Genet Dev. 2025 Aug;93:102376. doi: 10.1016/j.gde.2025.102376. Epub 2025 Jul 5. Curr Opin Genet Dev. 2025. PMID: 40617230 Review.
-
Heteroplasmy in action: tracking mtDNA segregation dynamics.EMBO J. 2024 Nov;43(22):5329-5331. doi: 10.1038/s44318-024-00226-x. Epub 2024 Sep 10. EMBO J. 2024. PMID: 39256559 Free PMC article.
References
-
- Alber AB, Paquet ER, Biserni M, Naef F, Suter DM (2018) Single live cell monitoring of protein turnover reveals intercellular variability and cell-cycle dependence of degradation rates. Mol Cell 71:1079–1091.e9 - PubMed
-
- Birky CW (1994) Relaxed and stringent genomes: why cytoplasmic genes don’t obey Mendel’s laws. J Heredity 85:355–365
-
- Birky J (2001) The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet 35:125–148 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

