Co-targeting SOS1 enhances the antitumor effects of KRASG12C inhibitors by addressing intrinsic and acquired resistance
- PMID: 39103541
- PMCID: PMC11424490
- DOI: 10.1038/s43018-024-00800-6
Co-targeting SOS1 enhances the antitumor effects of KRASG12C inhibitors by addressing intrinsic and acquired resistance
Abstract
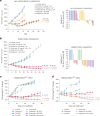
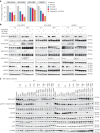
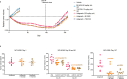
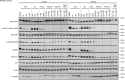
Combination approaches are needed to strengthen and extend the clinical response to KRASG12C inhibitors (KRASG12Ci). Here, we assessed the antitumor responses of KRASG12C mutant lung and colorectal cancer models to combination treatment with a SOS1 inhibitor (SOS1i), BI-3406, plus the KRASG12C inhibitor, adagrasib. We found that responses to BI-3406 plus adagrasib were stronger than to adagrasib alone, comparable to adagrasib with SHP2 (SHP2i) or EGFR inhibitors and correlated with stronger suppression of RAS-MAPK signaling. BI-3406 plus adagrasib treatment also delayed the emergence of acquired resistance and elicited antitumor responses from adagrasib-resistant models. Resistance to KRASG12Ci seemed to be driven by upregulation of MRAS activity, which both SOS1i and SHP2i were found to potently inhibit. Knockdown of SHOC2, a MRAS complex partner, partially restored response to KRASG12Ci treatment. These results suggest KRASG12C plus SOS1i to be a promising strategy for treating both KRASG12Ci naive and relapsed KRASG12C-mutant tumors.
© 2024. The Author(s).
Conflict of interest statement
V.T., S.J., K.K., D.A., H.A., O.B., K.B., D.G., M.G., M.H., S.L., A.J., C.A., T.M., G.M.-Z., P.A.J., R.L.S., S.S., F.T., M. Petronczki, N.K., M. Pearson, I.W. and M.H.H. report grants from the Austrian Research Promotion Agency (FFG) and have received personal fees from Boehringer Ingelheim (full-time employee) during the conduct of the study. V.T. is now a full-time employee for Exscientia, Vienna, Austria. M.H.H. and M.G. have been listed as the inventors on patent applications for SOS1 inhibitors. A.S., S.K., H.L., A.A.M., M.M., E.D.M., S.G., N.F., C.A.B., D.H.P., J.M.M., C.P.V., T.P.H. and J.R.M. report other from Boehringer Ingelheim (sponsored research) during the conduct of the study and this work was performed under a sponsored research collaboration between MD Anderson and Boehringer Ingelheim, for which the latter provided funding support. S. K. has ownership interest in Lutris, Iylon, Frontier Medicines, Xilis and Navire and is a consultant for Genentech, EMD Serono, Merck, Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, AstraZeneca/MedImmune, Bayer Health, Redx Pharma, Ipsen, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, Abbvie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics, Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical, Cardiff Oncology, Ono Pharmaceutical, Bristol-Myers Squibb-Medarex, Amgen, Tempus, Foundation Medicine, Harbinger Oncology, Takeda, CureTeq, Zentalis, Black Stone Therapeutics, NeoGenomics Laboratories and Accademia Nazionale Di Medicina, and receive research funding from Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly and Daiichi Sankyo. T.P.H. receives advisory fees from Cullgen and Roivant Discovery.
Figures





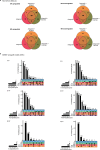
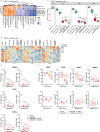

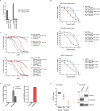
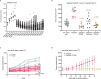
Update of
-
Combined KRASG12C and SOS1 inhibition enhances and extends the anti-tumor response in KRASG12C-driven cancers by addressing intrinsic and acquired resistance.bioRxiv [Preprint]. 2023 Jan 23:2023.01.23.525210. doi: 10.1101/2023.01.23.525210. bioRxiv. 2023. Update in: Nat Cancer. 2024 Sep;5(9):1352-1370. doi: 10.1038/s43018-024-00800-6. PMID: 36747713 Free PMC article. Updated. Preprint.
References
-
- Sacher, A. et al. Single-agent divarasib (GDC-6036) in solid tumors with a KRAS G12C mutation. N. Engl. J. Med.389, 710–721 (2023). - PubMed
-
- de Langen, A. J. et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: a randomised, open-label, phase 3 trial. Lancet401, 733–746 (2023). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- 892584/Österreichische Forschungsförderungsgesellschaft (Austrian Research Promotion Agency)
- P50 CA221707/CA/NCI NIH HHS/United States
- 883626/Österreichische Forschungsförderungsgesellschaft (Austrian Research Promotion Agency)
- 861507/Österreichische Forschungsförderungsgesellschaft (Austrian Research Promotion Agency)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

