Exome sequencing reveals neurodevelopmental genes in simplex consanguineous Iranian families with syndromic autism
- PMID: 39103847
- PMCID: PMC11302097
- DOI: 10.1186/s12920-024-01969-6
Exome sequencing reveals neurodevelopmental genes in simplex consanguineous Iranian families with syndromic autism
Abstract
Background and objective: Autosomal recessive genetic disorders pose significant health challenges in regions where consanguineous marriages are prevalent. The utilization of exome sequencing as a frequently employed methodology has enabled a clear delineation of diagnostic efficacy and mode of inheritance within multiplex consanguineous families. However, these aspects remain less elucidated within simplex families.
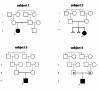
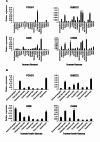
Methods: In this study involving 12 unrelated simplex Iranian families presenting syndromic autism, we conducted singleton exome sequencing. The identified genetic variants were validated using Sanger sequencing, and for the missense variants in FOXG1 and DMD, 3D protein structure modeling was carried out to substantiate their pathogenicity. To examine the expression patterns of the candidate genes in the fetal brain, adult brain, and muscle, RT-qPCR was employed.
Results: In four families, we detected an autosomal dominant gene (FOXG1), an autosomal recessive gene (CHKB), and two X-linked autism genes (IQSEC2 and DMD), indicating diverse inheritance patterns. In the remaining eight families, we were unable to identify any disease-associated genes. As a result, our variant detection rate stood at 33.3% (4/12), surpassing rates reported in similar studies of smaller cohorts. Among the four newly identified coding variants, three are de novo (heterozygous variant p.Trp546Ter in IQSEC2, heterozygous variant p.Ala188Glu in FOXG1, and hemizygous variant p.Leu211Met in DMD), while the homozygous variant p.Glu128Ter in CHKB was inherited from both healthy heterozygous parents. 3D protein structure modeling was carried out for the missense variants in FOXG1 and DMD, which predicted steric hindrance and spatial inhibition, respectively, supporting the pathogenicity of these human mutants. Additionally, the nonsense variant in CHKB is anticipated to influence its dimerization - crucial for choline kinase function - and the nonsense variant in IQSEC2 is predicted to eliminate three functional domains. Consequently, these distinct variants found in four unrelated individuals with autism are likely indicative of loss-of-function mutations.
Conclusions: In our two syndromic autism families, we discovered variants in two muscular dystrophy genes, DMD and CHKB. Given that DMD and CHKB are recognized for their participation in the non-cognitive manifestations of muscular dystrophy, it indicates that some genes transcend the boundary of apparently unrelated clinical categories, thereby establishing a novel connection between ASD and muscular dystrophy. Our findings also shed light on the complex inheritance patterns observed in Iranian consanguineous simplex families and emphasize the connection between autism spectrum disorder and muscular dystrophy. This underscores a likely genetic convergence between neurodevelopmental and neuromuscular disorders.
Keywords: De novo variants; Consanguineous simplex family; Exome sequencing; Iran; Muscular dystrophy; Neurodevelopmental disorder; Syndromic autism.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

