Hydrogen sulfide reduces oxidative stress in Huntington's disease via Nrf2
- PMID: 39104115
- PMCID: PMC11688542
- DOI: 10.4103/NRR.NRR-D-23-01051
Hydrogen sulfide reduces oxidative stress in Huntington's disease via Nrf2
Abstract
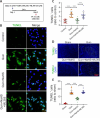
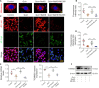
JOURNAL/nrgr/04.03/01300535-202506000-00028/figure1/v/2024-08-05T133530Z/r/image-tiff The pathophysiology of Huntington's disease involves high levels of the neurotoxin quinolinic acid. Quinolinic acid accumulation results in oxidative stress, which leads to neurotoxicity. However, the molecular and cellular mechanisms by which quinolinic acid contributes to Huntington's disease pathology remain unknown. In this study, we established in vitro and in vivo models of Huntington's disease by administering quinolinic acid to the PC12 neuronal cell line and the striatum of mice, respectively. We observed a decrease in the levels of hydrogen sulfide in both PC12 cells and mouse serum, which was accompanied by down-regulation of cystathionine β-synthase, an enzyme responsible for hydrogen sulfide production. However, treatment with NaHS (a hydrogen sulfide donor) increased hydrogen sulfide levels in the neurons and in mouse serum, as well as cystathionine β-synthase expression in the neurons and the mouse striatum, while also improving oxidative imbalance and mitochondrial dysfunction in PC12 cells and the mouse striatum. These beneficial effects correlated with upregulation of nuclear factor erythroid 2-related factor 2 expression. Finally, treatment with the nuclear factor erythroid 2-related factor 2 inhibitor ML385 reversed the beneficial impact of exogenous hydrogen sulfide on quinolinic acid-induced oxidative stress. Taken together, our findings show that hydrogen sulfide reduces oxidative stress in Huntington's disease by activating nuclear factor erythroid 2-related factor 2, suggesting that hydrogen sulfide is a novel neuroprotective drug candidate for treating patients with Huntington's disease.
Copyright © 2025 Copyright: © 2025 Neural Regeneration Research.
Conflict of interest statement
Figures





References
-
- Bains M, Kaur J, Akhtar A, Kuhad A, Sah SP. Anti-inflammatory effects of ellagic acid and vanillic acid against quinolinic acid-induced rat model of Huntington’s disease by targeting IKK-NF-κB pathway. Eur J Pharmacol. 2022;934:175316. - PubMed
-
- Bełtowski J. Hypoxia in the renal medulla: implications for hydrogen sulfide signaling. J Pharmacol Exp Ther. 2010;334:358–363. - PubMed
-
- Bhat A, Tan V, Heng B, Chow S, Basappa S, Essa MM, Chidambaram SB, Guillemin GJ. Papaverine, a phosphodiesterase 10A inhibitor, ameliorates quinolinic acid-induced synaptotoxicity in human cortical neurons. Neurotox Res. 2021;39:1238–1250. - PubMed
-
- Blomgren K, Hagberg H. Free radicals, mitochondria, and hypoxia-ischemia in the developing brain. Free Radic Biol Med. 2006;40:388–397. - PubMed
LinkOut - more resources
Full Text Sources

