Cytokine profile of anti-spike CD4+T cells predicts humoral and CD8+T cell responses after anti-SARS-CoV-2 mRNA vaccination
- PMID: 39104410
- PMCID: PMC11298648
- DOI: 10.1016/j.isci.2024.110441
Cytokine profile of anti-spike CD4+T cells predicts humoral and CD8+T cell responses after anti-SARS-CoV-2 mRNA vaccination
Abstract
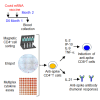
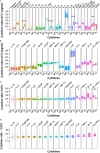
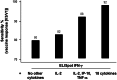
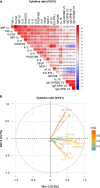
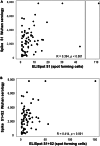
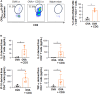
Coordinating immune responses - humoral and cellular - is vital for protection against severe Covid-19. Our study evaluates a multicytokine CD4+T cell signature's predictive for post-vaccinal serological and CD8+T cell responses. A cytokine signature composed of four cytokines (IL-2, TNF-α, IP10, IL-9) excluding IFN-γ, and generated through machine learning, effectively predicted the CD8+T cell response following mRNA-1273 or BNT162b2 vaccine administration. Its applicability extends to murine vaccination models, encompassing diverse immunization routes (such as intranasal) and vaccine platforms (including adjuvanted proteins). Notably, we found correlation between CD4+T lymphocyte-produced IL-21 and the humoral response. Consequently, we propose a test that offers a rapid overview of integrated immune responses. This approach holds particular relevance for scenarios involving immunocompromised patients because they often have low cell counts (lymphopenia) or pandemics. This study also underscores the pivotal role of CD4+T cells during a vaccine response and highlights their value in vaccine immunomonitoring.
Keywords: Health sciences; Immunity; Machine learning; Mathematical biosciences; Virology.
© 2024 The Authors.
Conflict of interest statement
DW is a consultant for Moderna. AS is a consultant for Gritstone Bio, Flow Pharma, Moderna, AstraZeneca, Qiagen, Fortress, Gilead, Sanofi, Merck, RiverVest, MedaCorp, Turnstone, NA Vaccine Institute, Emervax, Gerson Lehrman Group and Guggenheim. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work. OL participated in boards for Pfizer and Moderna. ET is consultant for Moderna and speakers for MSD and BMS.
Figures


References
-
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. - DOI - PMC - PubMed
-
- Oja A.E., Saris A., Ghandour C.A., Kragten N.A.M., Hogema B.M., Nossent E.J., Heunks L.M.A., Cuvalay S., Slot E., Linty F., et al. Divergent SARS-CoV-2-specific T- and B-cell responses in severe but not mild COVID-19 patients. Eur. J. Immunol. 2020;50:1998–2012. doi: 10.1002/eji.202048908. - DOI - PubMed
-
- Tarke A., Potesta M., Varchetta S., Fenoglio D., Iannetta M., Sarmati L., Mele D., Dentone C., Bassetti M., Montesano C., et al. Early and Polyantigenic CD4 T Cell Responses Correlate with Mild Disease in Acute COVID-19 Donors. Int. J. Mol. Sci. 2022;23:7155. doi: 10.3390/ijms23137155. - DOI - PMC - PubMed
-
- Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108728. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

