Sequence analysis of microbiota in clinical human cases with diabetic foot ulcers from China
- PMID: 39104504
- PMCID: PMC11298921
- DOI: 10.1016/j.heliyon.2024.e34368
Sequence analysis of microbiota in clinical human cases with diabetic foot ulcers from China
Abstract
Background: Diabetic foot ulcers (DFU) seriously threaten the health and quality of life of patients. The microbiota is the primary reason for the refractory and high recurrence of DFU. This study aimed to determine the wound microbiota at different DFU stages.
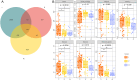
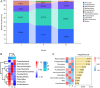
Methods: Wound samples were collected from 48 patients with DFU and divided into three phases: inflammatory (I, n = 49), proliferation (P, n = 22), and remodeling (R, n = 19). The wound samples obtained at different stages were then subjected to 16S rRNA gene sequencing. The number of operational taxonomic units (OTUs) in the different groups was calculated according to the criterion of 97 % sequence similarity. The diversity of the microbiota differentially presented bacterial taxa at the phylum and genus levels, and important phyla and genera in the different groups were further explored.
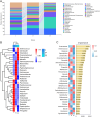
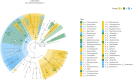
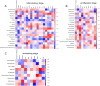
Results: After sequencing, 3351, 925, and 777 OTUs were observed in groups I, P, and R, respectively, and 175 OTUs overlapped. Compared with the inflammatory stage, the α-diversity of wound microbiota at proliferation and remodeling stages was significantly decreased (P < 0.05). At the phylum level, Firmicutes, Proteobacteria, Actinobacteriota, and Bacteroidota were the dominant phyla, accounting for more than 90 % of all the phyla. At the genus level, Random Forest and linear discriminant analysis effect size analyses showed that Peptoniphilus, Lactobacillus, Prevotella, Veillonella, Dialister, Streptococcus, and Ruminococcus were the signature wound microbiota for the inflammatory stage; Anaerococcus, Ralstonia, Actinomyces, and Akkermansia were important species for the proliferation stage; and the crucial genera for the remodeling stage were Enterobacter, Pseudomonas, Sondgrassella, Bifidobacterium, and Faecalibacterium.
Conclusions: There were significant differences in the composition and structure of the wound microbiota in patients with DFU at different stages, which may lay a foundation for effectively promoting wound healing in DFU.
Keywords: Diabetic foot ulcers; Inflammation stage; Proliferation stage; Remodeling stage; Wound microbiota.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Association Between the Diabetic Foot Ulcer and the Bacterial Colony of the Skin Based on 16S rRNA Gene Sequencing: An Observational Study.Clin Cosmet Investig Dermatol. 2023 Oct 10;16:2801-2812. doi: 10.2147/CCID.S425922. eCollection 2023. Clin Cosmet Investig Dermatol. 2023. PMID: 37841062 Free PMC article.
-
[Analysis of the dynamic changes in gut microbiota in patients with extremely severe burns by 16S ribosomal RNA high-throughput sequencing technology].Zhonghua Shao Shang Za Zhi. 2020 Dec 20;36(12):1159-1166. doi: 10.3760/cma.j.cn501120-20200518-00271. Zhonghua Shao Shang Za Zhi. 2020. PMID: 33379852 Chinese.
-
[Bacterial community diversity in Dermatophagoides farinae using high-throughput sequencing].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022 Nov 15;34(6):630-634. doi: 10.16250/j.32.1374.2022105. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022. PMID: 36642905 Chinese.
-
Gut Microbiota and Its Repercussion in Parkinson's Disease: A Systematic Review in Occidental Patients.Neurol Int. 2023 Jun 13;15(2):750-763. doi: 10.3390/neurolint15020047. Neurol Int. 2023. PMID: 37368331 Free PMC article. Review.
-
Current scenario of traditional medicines in management of diabetic foot ulcers: A review.World J Diabetes. 2023 Jan 15;14(1):1-16. doi: 10.4239/wjd.v14.i1.1. World J Diabetes. 2023. PMID: 36684382 Free PMC article. Review.
Cited by
-
The healing process of diabetic ulcers correlates with changes in the cutaneous microbiota.Sci Rep. 2024 Nov 12;14(1):27628. doi: 10.1038/s41598-024-77987-2. Sci Rep. 2024. PMID: 39528566 Free PMC article.
References
LinkOut - more resources
Full Text Sources

