FInCH: FIJI plugin for automated and scalable whole-image analysis of protein expression and cell morphology
- PMID: 39105087
- PMCID: PMC11299569
- DOI: 10.1016/j.mex.2024.102855
FInCH: FIJI plugin for automated and scalable whole-image analysis of protein expression and cell morphology
Abstract
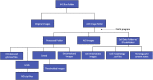
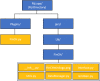
Study of morphogenesis and its regulation requires analytical tools that enable simultaneous assessment of processes operating at cellular level, such as synthesis of transcription factors (TF), with their effects at the tissue scale. Most current studies conduct histological, cellular and immunochemical (IHC) analyses in separate steps, introducing inevitable biases in finding and alignment of areas of interest at vastly distinct scales of organization, as well as image distortion associated with image repositioning or file modifications. These problems are particularly severe for longitudinal analyses of growing structures that change size and shape. Here we introduce a python-based application for automated and complete whole-slide measurement of expression of multiple TFs and associated cellular morphology. The plugin collects data at customizable scale from the cell-level to the entire structure, records each data point with positional information, accounts for ontogenetic transformation of structures and variation in slide positioning with scalable grid, and includes a customizable file manager that outputs collected data in association with full details of image classification (e.g., ontogenetic stage, population, IHC assay). We demonstrate the utility and accuracy of this application by automated measurement of morphology and associated expression of eight TFs for more than six million cells recorded with full positional information in beak tissues across 12 developmental stages and 25 study populations of a wild passerine bird. Our script is freely available as an open-source Fiji plugin and can be applied to IHC slides from any imaging platforms and transcriptional factors.
Keywords: Cell shape; Evolutionary diversification; Histology; Immunohistochemistry; Morphogenesis; Ontogeny; Scalable whole-image analysis of immunohistochemistry expression and cell segmentation: FInCH (File Iterating and Color deconvolving Histogram) plugin; Transcription factors.
© 2024 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







Similar articles
-
Whole-slide imaging and a Fiji-based image analysis workflow of immunohistochemistry staining of pancreatic islets.MethodsX. 2022 Sep 13;9:101856. doi: 10.1016/j.mex.2022.101856. eCollection 2022. MethodsX. 2022. PMID: 36204475 Free PMC article.
-
LSM-W2: laser scanning microscopy worker for wheat leaf surface morphology.BMC Syst Biol. 2019 Mar 5;13(Suppl 1):22. doi: 10.1186/s12918-019-0689-8. BMC Syst Biol. 2019. PMID: 30836965 Free PMC article.
-
IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples.PLoS One. 2014 May 6;9(5):e96801. doi: 10.1371/journal.pone.0096801. eCollection 2014. PLoS One. 2014. PMID: 24802416 Free PMC article.
-
AI (artificial intelligence) in histopathology--from image analysis to automated diagnosis.Folia Histochem Cytobiol. 2009 Jan;47(3):355-61. doi: 10.2478/v10042-009-0087-y. Folia Histochem Cytobiol. 2009. PMID: 20164018 Review.
-
Applications and challenges of digital pathology and whole slide imaging.Biotech Histochem. 2015 Jul;90(5):341-7. doi: 10.3109/10520295.2015.1044566. Epub 2015 May 15. Biotech Histochem. 2015. PMID: 25978139 Review.
References
-
- Wilkins A.S. Sinauer Associates; Sunderland, MA: 2001. The Evolution of Developmental Pathways.
-
- Kalyuzhny A.E. Springer; 2016. Immunohistochemistry: Essential Elements and Beyond.
-
- A.V. Badyaev, C.A. Lee, M.J. Gleason, G.A. Semenov, S.E. Britton, R.A. Duckworth, Tuning Regulatory Signaling Network to Cell Jamming Transitions can Delineate Population Divergence in Morphogenesis Under Review.
LinkOut - more resources
Full Text Sources
Miscellaneous

