Pseudomonas aeruginosa surface motility and invasion into competing communities enhance interspecies antagonism
- PMID: 39105585
- PMCID: PMC11389416
- DOI: 10.1128/mbio.00956-24
Pseudomonas aeruginosa surface motility and invasion into competing communities enhance interspecies antagonism
Abstract
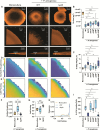
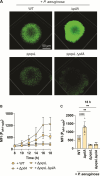
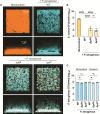
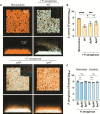
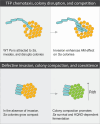
Chronic polymicrobial infections involving Pseudomonas aeruginosa and Staphylococcus aureus are prevalent, difficult to eradicate, and associated with poor health outcomes. Therefore, understanding interactions between these pathogens is important to inform improved treatment development. We previously demonstrated that P. aeruginosa is attracted to S. aureus using type IV pili (TFP)-mediated chemotaxis, but the impact of attraction on S. aureus growth and physiology remained unknown. Using live single-cell confocal imaging to visualize microcolony structure, spatial organization, and survival of S. aureus during coculture, we found that interspecies chemotaxis provides P. aeruginosa a competitive advantage by promoting invasion into and disruption of S. aureus microcolonies. This behavior renders S. aureus susceptible to P. aeruginosa antimicrobials. Conversely, in the absence of TFP motility, P. aeruginosa cells exhibit reduced invasion of S. aureus colonies. Instead, P. aeruginosa builds a cellular barrier adjacent to S. aureus and secretes diffusible, bacteriostatic antimicrobials like 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO) into the S. aureus colonies. Reduced invasion leads to the formation of denser and thicker S. aureus colonies with increased HQNO-mediated lactic acid fermentation, a physiological change that could complicate treatment strategies. Finally, we show that P. aeruginosa motility modifications of spatial structure enhance competition against S. aureus. Overall, these studies expand our understanding of how P. aeruginosa TFP-mediated interspecies chemotaxis facilitates polymicrobial interactions, highlighting the importance of spatial positioning in mixed-species communities.
Importance: The polymicrobial nature of many chronic infections makes their eradication challenging. Particularly, coisolation of Pseudomonas aeruginosa and Staphylococcus aureus from airways of people with cystic fibrosis and chronic wound infections is common and associated with severe clinical outcomes. The complex interplay between these pathogens is not fully understood, highlighting the need for continued research to improve management of chronic infections. Our study unveils that P. aeruginosa is attracted to S. aureus, invades into neighboring colonies, and secretes anti-staphylococcal factors into the interior of the colony. Upon inhibition of P. aeruginosa motility and thus invasion, S. aureus colony architecture changes dramatically, whereby S. aureus is protected from P. aeruginosa antagonism and responds through physiological alterations that may further hamper treatment. These studies reinforce accumulating evidence that spatial structuring can dictate community resilience and reveal that motility and chemotaxis are critical drivers of interspecies competition.
Keywords: Pseudomonas aeruginosa; Staphylococcus aureus; biofilms; polymicrobial; spatial organization; type IV pili motility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
Pseudomonas aeruginosa surface motility and invasion into competing communities enhances interspecies antagonism.bioRxiv [Preprint]. 2024 Apr 4:2024.04.03.588010. doi: 10.1101/2024.04.03.588010. bioRxiv. 2024. Update in: mBio. 2024 Sep 11;15(9):e0095624. doi: 10.1128/mbio.00956-24. PMID: 38617332 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R35 GM142760/GM/NIGMS NIH HHS/United States
- 2017879/National Science Foundation (NSF)
- RGY0077/2020/Human Frontier Science Program (HFSP)
- GAANN Fellowship
- AI007519/HHS | National Institutes of Health (NIH)
- R35GM142760/HHS | National Institutes of Health (NIH)
- 5T32HL007638-34/HHS | National Institutes of Health (NIH)
- R35 GM151158/GM/NIGMS NIH HHS/United States
- John H. Copenhaver Jr. Fellowship
- T32 AI007519/AI/NIAID NIH HHS/United States
- 1R35GM151158-01/HHS | National Institutes of Health (NIH)
- LIMOLI18F5,LIMOLI19R3/Cystic Fibrosis Foundation (CFF)
LinkOut - more resources
Full Text Sources

