1-Deoxy-d-xylulose 5-phosphate reductoisomerase as target for anti Toxoplasma gondii agents: crystal structure, biochemical characterization and biological evaluation of inhibitors
- PMID: 39105673
- PMCID: PMC11346426
- DOI: 10.1042/BCJ20240110
1-Deoxy-d-xylulose 5-phosphate reductoisomerase as target for anti Toxoplasma gondii agents: crystal structure, biochemical characterization and biological evaluation of inhibitors
Abstract
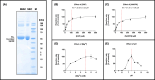
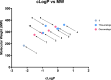
Toxoplasma gondii is a widely distributed apicomplexan parasite causing toxoplasmosis, a critical health issue for immunocompromised individuals and for congenitally infected foetuses. Current treatment options are limited in number and associated with severe side effects. Thus, novel anti-toxoplasma agents need to be identified and developed. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) is considered the rate-limiting enzyme in the non-mevalonate pathway for the biosynthesis of the isoprenoid precursors isopentenyl pyrophosphate and dimethylallyl pyrophosphate in the parasite, and has been previously investigated for its key role as a novel drug target in some species, encompassing Plasmodia, Mycobacteria and Escherichia coli. In this study, we present the first crystal structure of T. gondii DXR (TgDXR) in a tertiary complex with the inhibitor fosmidomycin and the cofactor NADPH in dimeric conformation at 2.5 Å resolution revealing the inhibitor binding mode. In addition, we biologically characterize reverse α-phenyl-β-thia and β-oxa fosmidomycin analogues and show that some derivatives are strong inhibitors of TgDXR which also, in contrast with fosmidomycin, inhibit the growth of T. gondii in vitro. Here, ((3,4-dichlorophenyl)((2-(hydroxy(methyl)amino)-2-oxoethyl)thio)methyl)phosphonic acid was identified as the most potent anti T. gondii compound. These findings will enable the future design and development of more potent anti-toxoplasma DXR inhibitors.
Keywords: Toxoplasma gondii; DXR; DXR inhibitors; SAXS; anti-infective; crystal structure; enzymatic assay; fosmidomycin; growth inhibition; parasite.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures









References
-
- Levine, N.D. (2018) The Protozoan Phylum Apicomplexa: Volume 2, CRC Press, Boca Raton, FL
-
- Centers for Disease Control and Prevention. Parasites - toxoplasmosis (toxoplasma infection) - epidemiology & risk factors, https://www.cdc.gov/parasites/toxoplasmosis/epi.html
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

