Central role of SUMOylation in the regulation of chromatin interactions and transcriptional outputs of the androgen receptor in prostate cancer cells
- PMID: 39106160
- PMCID: PMC11381344
- DOI: 10.1093/nar/gkae653
Central role of SUMOylation in the regulation of chromatin interactions and transcriptional outputs of the androgen receptor in prostate cancer cells
Abstract
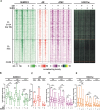
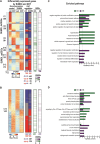
The androgen receptor (AR) is pivotal in prostate cancer (PCa) progression and represents a critical therapeutic target. AR-mediated gene regulation involves intricate interactions with nuclear proteins, with many mediating and undergoing post-translational modifications that present alternative therapeutic avenues. Through chromatin proteomics in PCa cells, we identified SUMO ligases together with nuclear receptor coregulators and pioneer transcription factors within the AR's protein network. Intriguingly, this network displayed a significant association with SUMO2/3. To elucidate the influence of SUMOylation on AR chromatin interactions and subsequent gene regulation, we inhibited SUMOylation using ML-792 (SUMOi). While androgens generally facilitated the co-occupancy of SUMO2/3 and AR on chromatin, SUMOi induced divergent effects dependent on the type of AR-binding site (ARB). SUMOi augmented AR's pioneer-like binding on inaccessible chromatin regions abundant in androgen response elements (AREs) and diminished its interaction with accessible chromatin regions sparse in AREs yet rich in pioneer transcription factor motifs. The SUMOi-impacted ARBs divergently influenced AR-regulated genes; those associated with AR-mediated activation played roles in negative regulation of cell proliferation, while those with AR-mediated repression were involved in pattern formation. In conclusion, our findings underscore the pervasive influence of SUMOylation in shaping AR's role in PCa cells, potentially unveiling new therapeutic strategies.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Shafi A.A., Yen A.E., Weigel N.L.. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 2013; 140:223–238. - PubMed
-
- Coffey K., Robson C.N.. Regulation of the androgen receptor by post-translational modifications. J. Endocrinol. 2012; 215:221–237. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

