Stress granule formation helps to mitigate neurodegeneration
- PMID: 39106168
- PMCID: PMC11381325
- DOI: 10.1093/nar/gkae655
Stress granule formation helps to mitigate neurodegeneration
Abstract
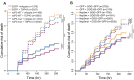
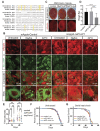
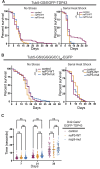
Cellular stress pathways that inhibit translation initiation lead to transient formation of cytoplasmic RNA/protein complexes known as stress granules. Many of the proteins found within stress granules and the dynamics of stress granule formation and dissolution are implicated in neurodegenerative disease. Whether stress granule formation is protective or harmful in neurodegenerative conditions is not known. To address this, we took advantage of the alphavirus protein nsP3, which selectively binds dimers of the central stress granule nucleator protein G3BP and markedly reduces stress granule formation without directly impacting the protein translational inhibitory pathways that trigger stress granule formation. In Drosophila and rodent neurons, reducing stress granule formation with nsP3 had modest impacts on lifespan even in the setting of serial stress pathway induction. In contrast, reducing stress granule formation in models of ataxia, amyotrophic lateral sclerosis and frontotemporal dementia largely exacerbated disease phenotypes. These data support a model whereby stress granules mitigate, rather than promote, neurodegenerative cascades.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Update of
-
Stress granule formation helps to mitigate neurodegeneration.bioRxiv [Preprint]. 2023 Nov 11:2023.11.07.566060. doi: 10.1101/2023.11.07.566060. bioRxiv. 2023. Update in: Nucleic Acids Res. 2024 Sep 9;52(16):9745-9759. doi: 10.1093/nar/gkae655. PMID: 37986813 Free PMC article. Updated. Preprint.
References
-
- Walter P., Ron D.. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011; 334:1081–1086. - PubMed
MeSH terms
Substances
Grants and funding
- R01 NS086810/NS/NINDS NIH HHS/United States
- F31 NS115257/NS/NINDS NIH HHS/United States
- 2018-03843/Swedish Research Council
- P50 HD104463/HD/NICHD NIH HHS/United States
- Ann Arbor Active Against ALS
- T32 NS076401/NS/NINDS NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- BLRD BX004842/Veterans Affairs
- I01 BX004842/BX/BLRD VA/United States
- P50HD104463/NH/NIH HHS/United States
- R01 NS099280/NS/NINDS NIH HHS/United States
- R01 NS113943/NS/NINDS NIH HHS/United States
- R01 NS097542/NS/NINDS NIH HHS/United States
- R56 NS128110/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

