Natural Killer Cell Regulation of Breast Cancer Stem Cells Mediates Metastatic Dormancy
- PMID: 39106452
- PMCID: PMC11474167
- DOI: 10.1158/0008-5472.CAN-24-0030
Natural Killer Cell Regulation of Breast Cancer Stem Cells Mediates Metastatic Dormancy
Abstract
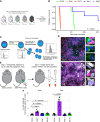
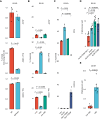
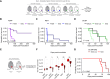
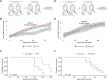
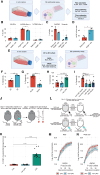
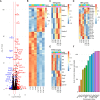
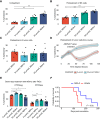
Patients with breast cancer with estrogen receptor-positive tumors face a constant risk of disease recurrence for the remainder of their lives. Dormant tumor cells residing in tissues such as the bone marrow may generate clinically significant metastases many years after initial diagnosis. Previous studies suggest that dormant cancer cells display "stem-like" properties (cancer stem cell, CSC), which may be regulated by the immune system. To elucidate the role of the immune system in controlling dormancy and its escape, we studied dormancy in immunocompetent, syngeneic mouse breast cancer models. Three mouse breast cancer cell lines, PyMT, Met1, and D2.0R, contained CSCs that displayed short- and long-term metastatic dormancy in vivo, which was dependent on the host immune system. Each model was regulated by different components of the immune system. Natural killer (NK) cells were key for the metastatic dormancy phenotype in D2.0R cells. Quiescent D2.0R CSCs were resistant to NK cell cytotoxicity, whereas proliferative CSCs were sensitive. Resistance to NK cell cytotoxicity was mediated, in part, by the expression of BACH1 and SOX2 transcription factors. Expression of STING and STING targets was decreased in quiescent CSCs, and the STING agonist MSA-2 enhanced NK cell killing. Collectively, these findings demonstrate the role of immune regulation of breast tumor dormancy and highlight the importance of utilizing immunocompetent models to study this phenomenon. Significance: The immune system controls disseminated breast cancer cells during disease latency, highlighting the need to utilize immunocompetent models to identify strategies for targeting dormant cancer cells and reducing metastatic recurrence. See related commentary by Cackowski and Korkaya, p. 3319.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
G.G. Bushnell reports grants from the National Institutes of Health during the conduct of the study. M. Burness reports grants from Pfizer and GSK during the conduct of the study and grants from Cyteir outside the submitted work. M.S. Wicha reports grants from NCI and Breast Cancer Research Foundation during the conduct of the study. No disclosures were reported by the other authors.
Figures

Update of
-
Natural killer cell regulation of breast cancer stem cells mediates metastatic dormancy.bioRxiv [Preprint]. 2023 Oct 3:2023.10.02.560493. doi: 10.1101/2023.10.02.560493. bioRxiv. 2023. Update in: Cancer Res. 2024 Oct 15;84(20):3337-3353. doi: 10.1158/0008-5472.CAN-24-0030. PMID: 37873211 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

