Expanding the genetic and phenotypic landscape of replication factor C complex-related disorders: RFC4 deficiency is linked to a multisystemic disorder
- PMID: 39106866
- PMCID: PMC11393705
- DOI: 10.1016/j.ajhg.2024.07.008
Expanding the genetic and phenotypic landscape of replication factor C complex-related disorders: RFC4 deficiency is linked to a multisystemic disorder
Abstract
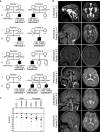
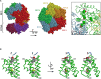
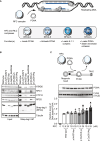
The precise regulation of DNA replication is vital for cellular division and genomic integrity. Central to this process is the replication factor C (RFC) complex, encompassing five subunits, which loads proliferating cell nuclear antigen onto DNA to facilitate the recruitment of replication and repair proteins and enhance DNA polymerase processivity. While RFC1's role in cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS) is known, the contributions of RFC2-5 subunits on human Mendelian disorders is largely unexplored. Our research links bi-allelic variants in RFC4, encoding a core RFC complex subunit, to an undiagnosed disorder characterized by incoordination and muscle weakness, hearing impairment, and decreased body weight. We discovered across nine affected individuals rare, conserved, predicted pathogenic variants in RFC4, all likely to disrupt the C-terminal domain indispensable for RFC complex formation. Analysis of a previously determined cryo-EM structure of RFC bound to proliferating cell nuclear antigen suggested that the variants disrupt interactions within RFC4 and/or destabilize the RFC complex. Cellular studies using RFC4-deficient HeLa cells and primary fibroblasts demonstrated decreased RFC4 protein, compromised stability of the other RFC complex subunits, and perturbed RFC complex formation. Additionally, functional studies of the RFC4 variants affirmed diminished RFC complex formation, and cell cycle studies suggested perturbation of DNA replication and cell cycle progression. Our integrated approach of combining in silico, structural, cellular, and functional analyses establishes compelling evidence that bi-allelic loss-of-function RFC4 variants contribute to the pathogenesis of this multisystemic disorder. These insights broaden our understanding of the RFC complex and its role in human health and disease.
Keywords: DNA replication; Mendelian disorder; gene discovery; rare disease; replication factor C complex; translational research.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Guilliam T.A., Yeeles J.T.P. An updated perspective on the polymerase division of labor during eukaryotic DNA replication. Crit. Rev. Biochem. Mol. Biol. 2020;55:469–481. - PubMed
-
- Bernad A., Blanco L., Lazaro J.M., Martin G., Salas M. A conserved 3'→5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. - PubMed
-
- Shevelev I.V., Hubscher U. The 3'–5' exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. - PubMed
-
- Bellelli R., Boulton S.J. Spotlight on the Replisome: aetiology of DNA replication-associated genetic diseases. Trends Genet. 2021;37:317–336. - PubMed
-
- Schmidt S.L., Pautz A.L., Burgers P.M. ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J. Biol. Chem. 2001;276:34792–34800. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

