HDAC3 genetic and pharmacologic inhibition radiosensitizes fusion positive rhabdomyosarcoma by promoting DNA double-strand breaks
- PMID: 39107280
- PMCID: PMC11303816
- DOI: 10.1038/s41420-024-02115-y
HDAC3 genetic and pharmacologic inhibition radiosensitizes fusion positive rhabdomyosarcoma by promoting DNA double-strand breaks
Abstract
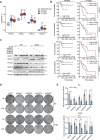
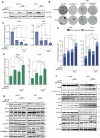
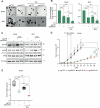
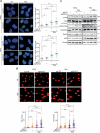
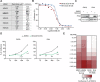
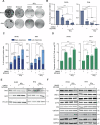
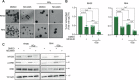
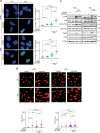
Radiotherapy (RT) plays a critical role in the management of rhabdomyosarcoma (RMS), the prevalent soft tissue sarcoma in childhood. The high risk PAX3-FOXO1 fusion-positive subtype (FP-RMS) is often resistant to RT. We have recently demonstrated that inhibition of class-I histone deacetylases (HDACs) radiosensitizes FP-RMS both in vitro and in vivo. However, HDAC inhibitors exhibited limited success on solid tumors in human clinical trials, at least in part due to the presence of off-target effects. Hence, identifying specific HDAC isoforms that can be targeted to radiosensitize FP-RMS is imperative. We, here, found that only HDAC3 silencing, among all class-I HDACs screened by siRNA, radiosensitizes FP-RMS cells by inhibiting colony formation. Thus, we dissected the effects of HDAC3 depletion using CRISPR/Cas9-dependent HDAC3 knock-out (KO) in FP-RMS cells, which resulted in Endoplasmatic Reticulum Stress activation, ERK inactivation, PARP1- and caspase-dependent apoptosis and reduced stemness when combined with irradiation compared to single treatments. HDAC3 loss-of-function increased DNA damage in irradiated cells augmenting H2AX phosphorylation and DNA double-strand breaks (DSBs) and counteracting irradiation-dependent activation of ATM and DNA-Pkcs as well as Rad51 protein induction. Moreover, HDAC3 depletion hampers FP-RMS tumor growth in vivo and maximally inhibits the growth of irradiated tumors compared to single approaches. We, then, developed a new HDAC3 inhibitor, MC4448, which showed specific cell anti-tumor effects and mirrors the radiosensitizing effects of HDAC3 depletion in vitro synergizing with ERKs inhibition. Overall, our findings dissect the pro-survival role of HDAC3 in FP-RMS and suggest HDAC3 genetic or pharmacologic inhibition as a new promising strategy to overcome radioresistance in this tumor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Yechieli RL, Mandeville HC, Hiniker SM, Bernier‐Chastagner V, McGovern S, Scarzello G, et al. Rhabdomyosarcoma. Pediatr Blood Cancer. 2021; 68. 10.1002/pbc.28254. - PubMed
Grants and funding
- IG #15312/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- GR-2016-02364546/Ministero della Salute (Ministry of Health, Italy)
- RG120172B8E53D03/Sapienza Università di Roma (Sapienza University of Rome)
- IG26172/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- GR-2021-12374415/Ministero della Salute (Ministry of Health, Italy)
- IG #10338/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- P30 CA045508/CA/NCI NIH HHS/United States
- IG #24696/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- GR-2013 02359212/Ministero della Salute (Ministry of Health, Italy)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

