53BP1 loss elicits cGAS-STING-dependent antitumor immunity in ovarian and pancreatic cancer
- PMID: 39107288
- PMCID: PMC11303708
- DOI: 10.1038/s41467-024-50999-2
53BP1 loss elicits cGAS-STING-dependent antitumor immunity in ovarian and pancreatic cancer
Abstract
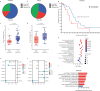
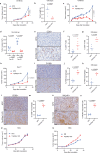
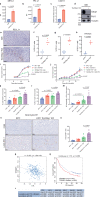
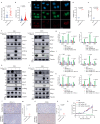
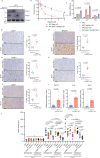
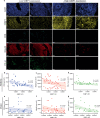
53BP1 nucleates the anti-end resection machinery at DNA double-strand breaks, thereby countering BRCA1 activity. Loss of 53BP1 leads to DNA end processing and homologous recombination in BRCA1-deficient cells. Consequently, BRCA1-mutant tumors, typically sensitive to PARP inhibitors (PARPi), become resistant in the absence of 53BP1. Here, we demonstrate that the 'leaky' DNA end resection in the absence of 53BP1 results in increased micronuclei and cytoplasmic double-stranded DNA, leading to activation of the cGAS-STING pathway and pro-inflammatory signaling. This enhances CD8+ T cell infiltration, activates macrophages and natural killer cells, and impedes tumor growth. Loss of 53BP1 correlates with a response to immune checkpoint blockade (ICB) and improved overall survival. Immunohistochemical assessment of 53BP1 in two malignancies, high grade serous ovarian cancer and pancreatic ductal adenocarcinoma, which are refractory to ICBs, reveals that lower 53BP1 levels correlate with an increased adaptive and innate immune response. Finally, BRCA1-deficient tumors that develop resistance to PARPi due to the loss of 53BP1 are susceptible to ICB. Therefore, we conclude that 53BP1 is critical for tumor immunogenicity and underpins the response to ICB. Our results support including 53BP1 expression as an exploratory biomarker in ICB trials for malignancies typically refractory to immunotherapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
- R01 CA208244/CA/NCI NIH HHS/United States
- W81XWH-15-0564/OC140632/U.S. Department of Defense (United States Department of Defense)
- R01 CA264900/CA/NCI NIH HHS/United States
- CA208244/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA248306/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

