eIF4E-independent translation is largely eIF3d-dependent
- PMID: 39107322
- PMCID: PMC11303786
- DOI: 10.1038/s41467-024-51027-z
eIF4E-independent translation is largely eIF3d-dependent
Abstract
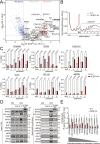
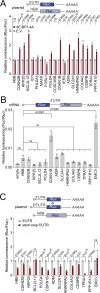
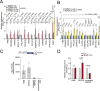
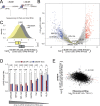
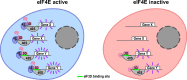
Translation initiation is a highly regulated step needed for protein synthesis. Most cell-based mechanistic work on translation initiation has been done using non-stressed cells growing in medium with sufficient nutrients and oxygen. This has yielded our current understanding of 'canonical' translation initiation, involving recognition of the mRNA cap by eIF4E1 followed by successive recruitment of initiation factors and the ribosome. Many cells, however, such as tumor cells, are exposed to stresses such as hypoxia, low nutrients or proteotoxic stress. This leads to inactivation of mTORC1 and thereby inactivation of eIF4E1. Hence the question arises how cells translate mRNAs under such stress conditions. We study here how mRNAs are translated in an eIF4E1-independent manner by blocking eIF4E1 using a constitutively active version of eIF4E-binding protein (4E-BP). Via ribosome profiling we identify a subset of mRNAs that are still efficiently translated when eIF4E1 is inactive. We find that these mRNAs preferentially release eIF4E1 when eIF4E1 is inactive and bind instead to eIF3d via its cap-binding pocket. eIF3d then enables these mRNAs to be efficiently translated due to its cap-binding activity. In sum, our work identifies eIF3d-dependent translation as a major mechanism enabling mRNA translation in an eIF4E-independent manner.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

