High production of recombinant protein using geminivirus-based deconstructed vectors in Nicotiana benthamiana
- PMID: 39109056
- PMCID: PMC11300340
- DOI: 10.3389/fpls.2024.1407240
High production of recombinant protein using geminivirus-based deconstructed vectors in Nicotiana benthamiana
Abstract
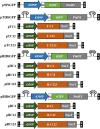
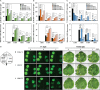
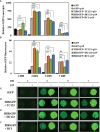
We focused on the geminiviral vector systems to develop an efficient vector system for plant biotechnology. Begomoviruses and curtoviruses, which belong to the Geminiviridae family, contain an intergenic region (IR) and four genes involved in replication, including replication-associated protein (Rep, C1), transcriptional activator (TrAP, C2), and replication enhancer (REn, C3). Geminiviruses can amplify thousands of copies of viral DNA using plant DNA polymerase and viral replication-related enzymes and accumulate viral proteins at high concentrations. In this study, we optimized geminiviral DNA replicon vectors based on tomato yellow leaf curl virus (TYLCV), honeysuckle yellow vein virus (HYVV), and mild curly top virus (BMCTV) for the rapid, high-yield plant-based production of recombinant proteins. Confirmation of the optimal combination by co-delivery of each replication-related gene and each IR harboring the Pontellina plumata-derived turbo green fluorescence protein (tGFP) gene via agroinfiltration in Nicotiana benthamiana leaves resulted in efficient replicon amplification and robust protein production within 3 days. Co-expression with the p19 protein of the tomato bush stunt virus, a gene-silencing suppressor, further enhanced tGFP accumulation by stabilizing mRNA. With this system, tGFP protein was produced at 0.7-1.2 mg/g leaf fresh weight, corresponding to 6.9-12.1% in total soluble protein. These results demonstrate the advantages of rapid and high-level production of recombinant proteins using the geminiviral DNA replicon system for transient expression in plants.
Keywords: Nicotiana benthamiana; geminivirus; transient expression; turbo green fluorescence protein; viral vector.
Copyright © 2024 Kim, Lee, Lee, Kil, Lee and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A DNA replicon system for rapid high-level production of virus-like particles in plants.Biotechnol Bioeng. 2009 Jul 1;103(4):706-14. doi: 10.1002/bit.22299. Biotechnol Bioeng. 2009. PMID: 19309755 Free PMC article.
-
Transcriptional reprogramming caused by the geminivirus Tomato yellow leaf curl virus in local or systemic infections in Nicotiana benthamiana.BMC Genomics. 2019 Jul 4;20(1):542. doi: 10.1186/s12864-019-5842-7. BMC Genomics. 2019. PMID: 31272383 Free PMC article.
-
A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication.J Virol. 2006 Apr;80(7):3624-33. doi: 10.1128/JVI.80.7.3624-3633.2006. J Virol. 2006. PMID: 16537630 Free PMC article.
-
High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system.Biotechnol Bioeng. 2010 May 1;106(1):9-17. doi: 10.1002/bit.22652. Biotechnol Bioeng. 2010. PMID: 20047189 Free PMC article.
-
Discovering host genes involved in the infection by the Tomato Yellow Leaf Curl Virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing.Viruses. 2013 Mar 22;5(3):998-1022. doi: 10.3390/v5030998. Viruses. 2013. PMID: 23524390 Free PMC article. Review.
Cited by
-
Bioluminescence-Driven Optimization of Geminivirus-Based Vectors as Tools for Plant Biotechnology.ACS Synth Biol. 2025 Aug 15;14(8):3078-3090. doi: 10.1021/acssynbio.5c00164. Epub 2025 Aug 4. ACS Synth Biol. 2025. PMID: 40758859 Free PMC article.
-
Expression of kiwifruit-derived actinidin in Nicotiana benthamiana leaves.Front Plant Sci. 2025 Jan 10;15:1532170. doi: 10.3389/fpls.2024.1532170. eCollection 2024. Front Plant Sci. 2025. PMID: 39866318 Free PMC article.
-
High-Level Production of a Recombinant Protein in Nicotiana benthamiana Leaves Through Transient Expression Using a Double Terminator.Int J Mol Sci. 2024 Oct 28;25(21):11573. doi: 10.3390/ijms252111573. Int J Mol Sci. 2024. PMID: 39519125 Free PMC article.
-
Virus-Induced Genome Editing (VIGE): One Step Away from an Agricultural Revolution.Int J Mol Sci. 2025 May 11;26(10):4599. doi: 10.3390/ijms26104599. Int J Mol Sci. 2025. PMID: 40429744 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

