Colorectal Cancer Recurrence Prediction Using a Tissue-Free Epigenomic Minimal Residual Disease Assay
- PMID: 39110016
- PMCID: PMC11443202
- DOI: 10.1158/1078-0432.CCR-24-1651
Colorectal Cancer Recurrence Prediction Using a Tissue-Free Epigenomic Minimal Residual Disease Assay
Abstract
Purpose: Posttreatment detection of ctDNA is strongly predictive of recurrence. Most minimal/molecular residual disease assays require prior tissue testing to guide ctDNA analysis, resulting in lengthy time to initial results and unevaluable patients.
Experimental design: We assessed a tissue-free assay (Guardant Reveal) that bioinformatically evaluates >20,000 epigenomic regions for ctDNA detection in 1,977 longitudinally collected postoperative plasma samples from 342 patients with resected colorectal cancer.
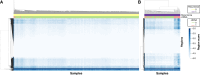
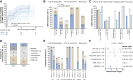
Results: We observed sensitive and specific detection of minimal/molecular residual disease associated with clinically meaningful differences in recurrence-free intervals at each time point evaluated with a median lead time of 5.3 months. The longitudinal sensitivity in stage II or higher colon cancer was 81%. Sensitivity increased with serial measurement and varied by recurrence site: higher for liver (100%) versus lung (53%) and peritoneal (40%). Sensitivity among patients with rectal cancer was 60% owing to a high proportion of lung metastases. Specificity was 98.2% among 1,461 posttreatment samples (99.1% among those with follow-up longer than the upper IQR of the lead time observed in this study).
Conclusions: Our data demonstrate the potential clinical utility of ctDNA as a tool to improve the management of stage II and higher colorectal cancer with a methodology that is noninvasive, accessible, and allows for rapid evaluation to inform clinical decisions.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
Y. Nakamura reports personal fees from Guardant Health Pte Ltd and Guardant Health Japan Corp, grants from Guardant Health AMEA, Inc and Guardant Health during the conduct of the study, personal fees from Natera, Inc, Roche Ltd, Premo Partners, Inc, Takeda, Exact Sciences, Gilead Sciences, MSD K.K., Eisai, Zeria Pharmaceutical, Miyarisan Pharmaceutical, Merck, Carenet, Inc, Hisamitsu Pharmaceutical, Taiho Pharmaceutical, Becton, Dickinson and Company; grants and personal fees from Seagen, Inc, Daiichi Sankyo Co Ltd, and Chugai Pharmaceutical; and grants from Genomedia, Tempus, and Roche Diagnostics K.K. outside the submitted work. N. Matsuhashi reports grants and personal fees from Abbott, Asahi Kasei Pharma, Chugai Pharmaceutical, Covidien Japan, Eli Lilly Japan, Eisai, Johnson & Johnson, Kaken Pharmaceutical, Kyowa Kirin, Terumo, and Tsumura; personal fees from AstraZeneca, Bayer Yakuhin, Bristol Myers Squibb, EA Pharma, Gunze Medical Limited, MC Medical, Merck BioPharma Japan, Miyarisan Pharmaceutical, Novartis, Olympus Marketing, Inc, Stryker, Takeda Pharmaceuticals, Viatris, and Yakult Honsha; grants, personal fees, and other support from Daiichi Sankyo; other support from EP‐CRSU, EPS Corporation, and ShiftZero K.K.; personal fees and other support from MSD and Ono Pharmaceutical; grants from Nippon Kayaku, Otsuka Pharmaceutical, and Toray Medical; and grants, personal fees, and nonfinancial support from Taiho Pharmaceutical outside the submitted work. E. Oki reports personal fees from Chugai Pharmaceutical, Bristol Meyers Squibb, Ono Pharmaceutical, Eli Lilly and Company, and Takeda Pharmaceutical and grants from Guardant Health, Inc outside the submitted work. M. Goto reports personal fees from Tsumura & Co, Daiichi Sankyo Company, Limited, Ono Pharmaceutical Co Ltd, and MSD K.K.; grants and personal fees from Taiho Pharmaceutical; and grants from Chugai Pharmaceutical and Nippon Kayaku outside the submitted work. Y. Kagawa reports personal fees from Chugai, Taiho, Ono, Merck, Takeda, and MSD outside the submitted work. T. Ohta reports personal fees from Bristol Myers Squibb Japan, Novartis AG, Daiichi Sankyo Company, Limited, EA Pharma Co, Ltd, Eli Lilly Japan K.K., Merck & Co, MSD K.K., Ono Pharmaceutical Co, Ltd, Otsuka Pharmaceutical Co, Ltd, Taiho Pharmaceutical Co, Ltd, Takeda Pharmaceutical Company Limited, AstraZeneca, and Yakult Honsha outside the submitted work. H. Bando reports personal fees from Eli Lilly Japan, Taiho Pharmaceutical, and Ono Pharmaceutical outside the submitted work. T. Yoshino reports grants and personal fees from Chugai Pharmaceutical, Takeda Pharmaceutical, Ono Pharmaceutical, and MSD K.K.; personal fees from Merck Biopharma, Bayer Yakuhin, and Sumitomo Corp; and grants from Amgen, Bristol Myers Squibb, Daiichi Sankyo, Eisai, Falco Biosystems, Genomedia, Medical & Biological Laboratories, Merus N.V., Molecular Health GmbH, Nippon Boehringer Ingelheim, Pfizer, Roche Diagnostics, Sanofi, Sysmex, and Taiho Pharmaceutical outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
-
- André T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol 2009;27:3109–16. - PubMed
-
- Yoshino T, Oki E, Misumi T, Kotaka M, Manaka D, Eto T, et al. Final analysis of 3 versus 6 months of adjuvant oxaliplatin and fluoropyrimidine-based therapy in patients with stage III colon cancer: the randomized phase III ACHIEVE trial. J Clin Oncol 2022;40:3419–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

