DNA hypo-methylation and expression of GBP4 induces T cell exhaustion in pancreatic cancer
- PMID: 39110249
- PMCID: PMC11306721
- DOI: 10.1007/s00262-024-03786-3
DNA hypo-methylation and expression of GBP4 induces T cell exhaustion in pancreatic cancer
Abstract
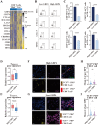
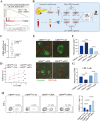
Immunotherapy for pancreatic ductal carcinoma (PDAC) remains disappointing due to the repressive tumor microenvironment and T cell exhaustion, in which the roles of interferon-stimulated genes were largely unknown. Here, we focused on a typical interferon-stimulated gene, GBP4, and investigated its potential diagnostic and therapeutic value in pancreatic cancer. Expression analysis on both local samples and public databases indicated that GBP4 was one of the most dominant GBP family members present in the PDAC microenvironment, and the expression level of GBP4 was negatively associated with patient survival. We then identified DNA hypo-methylation in regulatory regions of GBP4 in PDAC, and validated its regulatory role on GBP4 expression via performing targeted methylation using dCas9-SunTag-DNMAT3A-sgRNA-targeted methylation system on selected DNA locus. After that, we investigated the downstream functions of GBP4, and chemotaxis assays indicated that GBP4 overexpression significantly improved the infiltration of CD8+T cells, but also induced upregulation of immune checkpoint genes and T cell exhaustion. Lastly, in vitro T cell killing assays using primary organoids suggested that the PDAC samples with high level of GBP4 expression displayed significantly higher sensitivity to anti-PD-1 treatment. Taken together, our studies revealed the expression patterns and epigenetic regulatory mechanisms of GBP4 in pancreatic cancer and clarified the effects of GBP4 on T cell exhaustion and antitumor immunology.
Keywords: DNA methylation; GBP4; PD-1 blockade; Pancreatic ductal adenocarcinoma; T cell exhaustion.
© 2024. The Author(s).
Conflict of interest statement
All authors contributed to the article and approved the submitted version, and no conflict of interest exists in the submission of this manuscript.
Figures





Similar articles
-
The TMBIM1-YBX1 axis orchestrates MDSC recruitment and immunosuppressive microenvironment in pancreatic cancer.Theranostics. 2025 Feb 3;15(7):2794-2813. doi: 10.7150/thno.111180. eCollection 2025. Theranostics. 2025. PMID: 40083936 Free PMC article.
-
Single-cell RNA sequencing reveals compartmental remodeling of tumor-infiltrating immune cells induced by anti-CD47 targeting in pancreatic cancer.J Hematol Oncol. 2019 Nov 27;12(1):124. doi: 10.1186/s13045-019-0822-6. J Hematol Oncol. 2019. PMID: 31771616 Free PMC article.
-
Elevated SAMD3 expression in T cells predicts improved survival in pancreatic ductal adenocarcinoma patients.Cancer Immunol Immunother. 2025 Feb 1;74(3):93. doi: 10.1007/s00262-025-03948-x. Cancer Immunol Immunother. 2025. PMID: 39891760 Free PMC article.
-
Analysis of PD-L1 promoter methylation combined with immunogenic context in pancreatic ductal adenocarcinoma.Cancer Immunol Immunother. 2024 Jun 4;73(8):149. doi: 10.1007/s00262-024-03745-y. Cancer Immunol Immunother. 2024. PMID: 38833018 Free PMC article.
-
Immune Checkpoint Inhibition for Pancreatic Ductal Adenocarcinoma: Current Limitations and Future Options.Front Immunol. 2018 Aug 15;9:1878. doi: 10.3389/fimmu.2018.01878. eCollection 2018. Front Immunol. 2018. PMID: 30158932 Free PMC article. Review.
Cited by
-
Novel Insights into T-Cell Exhaustion and Cancer Biomarkers in PDAC Using ScRNA-Seq.Biology (Basel). 2025 Aug 7;14(8):1015. doi: 10.3390/biology14081015. Biology (Basel). 2025. PMID: 40906153 Free PMC article.
-
Multifaceted Roles of Guanylate-Binding Proteins in Cancer.Int J Mol Sci. 2025 Jun 7;26(12):5477. doi: 10.3390/ijms26125477. Int J Mol Sci. 2025. PMID: 40564939 Free PMC article. Review.
-
Postnatal epigenetic differences in calves following transient fetal infection with bovine viral diarrhea virus.BMC Genomics. 2025 May 2;26(1):441. doi: 10.1186/s12864-025-11562-5. BMC Genomics. 2025. PMID: 40316897 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

