Merkel cell polyomavirus small tumor antigen contributes to immune evasion by interfering with type I interferon signaling
- PMID: 39110744
- PMCID: PMC11333005
- DOI: 10.1371/journal.ppat.1012426
Merkel cell polyomavirus small tumor antigen contributes to immune evasion by interfering with type I interferon signaling
Abstract
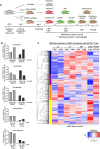
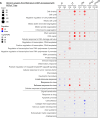
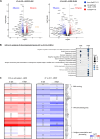
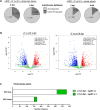
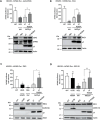
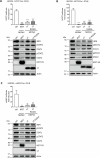
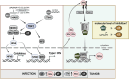
Merkel cell polyomavirus (MCPyV) is the causative agent of the majority of Merkel cell carcinomas (MCC). The virus has limited coding capacity, with its early viral proteins, large T (LT) and small T (sT), being multifunctional and contributing to infection and transformation. A fundamental difference in early viral gene expression between infection and MCPyV-driven tumorigenesis is the expression of a truncated LT (LTtr) in the tumor. In contrast, sT is expressed in both conditions and contributes significantly to oncogenesis. Here, we identified novel functions of early viral proteins by performing genome-wide transcriptome and chromatin studies in primary human fibroblasts. Due to current limitations in infection and tumorigenesis models, we mimic these conditions by ectopically expressing sT, LT or LTtr, individually or in combination, at different time points. In addition to its known function in cell cycle and inflammation modulation, we reveal a fundamentally new function of sT. We show that sT regulates the type I interferon (IFN) response downstream of the type I interferon receptor (IFNAR) by interfering with the interferon-stimulated gene factor 3 (ISGF3)-induced interferon-stimulated gene (ISG) response. Expression of sT leads to a reduction in the expression of interferon regulatory factor 9 (IRF9) which is a central component of the ISGF3 complex. We further show that this function of sT is conserved in BKPyV. We provide a first mechanistic understanding of which early viral proteins trigger and control the type I IFN response, which may influence MCPyV infection, persistence and, during MCC progression, regulation of the tumor microenvironment.
Copyright: © 2024 Ohnezeit et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Neumann F, Borchert S, Schmidt C, Reimer R, Hohenberg H, Fischer N, et al. Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome. PLoS One. 2011;6(12):e29112. doi: 10.1371/journal.pone.0029112 ; PubMed Central PMCID: PMC3246459. - DOI - PMC - PubMed
-
- Theiss JM, Gunther T, Alawi M, Neumann F, Tessmer U, Fischer N, et al. A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence. PLoS Pathog. 2015;11(7):e1004974. doi: 10.1371/journal.ppat.1004974 ; PubMed Central PMCID: PMC4517807. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

