Cell-specific cross-talk proteomics reveals cathepsin B signaling as a driver of glioblastoma malignancy near the subventricular zone
- PMID: 39110807
- PMCID: PMC11305394
- DOI: 10.1126/sciadv.adn1607
Cell-specific cross-talk proteomics reveals cathepsin B signaling as a driver of glioblastoma malignancy near the subventricular zone
Abstract
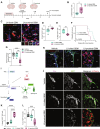
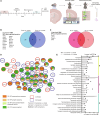
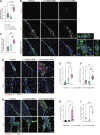
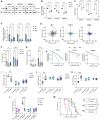
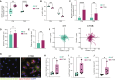
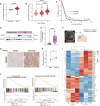
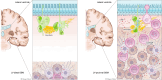
Glioblastoma (GBM) is the most prevalent and aggressive malignant primary brain tumor. GBM proximal to the lateral ventricles (LVs) is more aggressive, potentially because of subventricular zone contact. Despite this, cross-talk between GBM and neural stem/progenitor cells (NSC/NPCs) is not well understood. Using cell-specific proteomics, we show that LV-proximal GBM prevents neuronal maturation of NSCs through induction of senescence. In addition, GBM brain tumor-initiating cells (BTICs) increase expression of cathepsin B (CTSB) upon interaction with NPCs. Lentiviral knockdown and recombinant protein experiments reveal that both cell-intrinsic and soluble CTSB promote malignancy-associated phenotypes in BTICs. Soluble CTSB stalls neuronal maturation in NPCs while promoting senescence, providing a link between LV-tumor proximity and neurogenesis disruption. Last, we show LV-proximal CTSB up-regulation in patients, showing the relevance of this cross-talk in human GBM biology. These results demonstrate the value of proteomic analysis in tumor microenvironment research and provide direction for new therapeutic strategies in GBM.
Figures

Update of
-
Cell-specific crosstalk proteomics reveals cathepsin B signaling as a driver of glioblastoma malignancy near the subventricular zone.bioRxiv [Preprint]. 2023 Aug 21:2023.08.19.553966. doi: 10.1101/2023.08.19.553966. bioRxiv. 2023. Update in: Sci Adv. 2024 Aug 9;10(32):eadn1607. doi: 10.1126/sciadv.adn1607. PMID: 37662251 Free PMC article. Updated. Preprint.
References
-
- Bartkowiak T., Lima S. M., Hayes M. J., Mistry A. M., Brockman A. A., Sinnaeve J., Leelatian N., Roe C. E., Mobley B. C., Chotai S., Weaver K. D., Thompson R. C., Chambless L. B., Ihrie R. A., Irish J. M., An immunosuppressed microenvironment distinguishes lateral ventricle-contacting glioblastomas. JCI Insight 8, e160652 (2023). - PMC - PubMed
-
- Chaichana K. L., McGirt M. J., Frazier J., Attenello F., Guerrero-Cazares H., Quinones-Hinojosa A., Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J. Neurooncol. 89, 219–224 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

