Skin hepcidin initiates psoriasiform skin inflammation via Fe-driven hyperproliferation and neutrophil recruitment
- PMID: 39112467
- PMCID: PMC11306357
- DOI: 10.1038/s41467-024-50993-8
Skin hepcidin initiates psoriasiform skin inflammation via Fe-driven hyperproliferation and neutrophil recruitment
Abstract
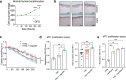
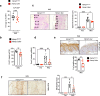
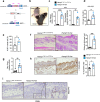
Psoriasis is a multifactorial, chronic inflammatory skin disease with unresolved questions on its primary events. Iron overload has been described in the epidermis of psoriasis patients, but its relevance remains unknown. We found that the key iron regulatory hormone hepcidin was highly expressed in the epidermis of psoriasis patients, especially the pustular variants resistant to treatments. In a murine model of acute skin inflammation, keratinocyte-derived hepcidin was required for iron retention in keratinocytes, leading to hyperproliferation of the epidermal layer and neutrophil recruitment, two main features of psoriatic skin lesions. Keratinocytes overexpressing hepcidin were sufficient to elicit these psoriasiform features in a transgenic mouse model. Furthermore, transcriptome analysis of these keratinocytes revealed canonical pathways found in human psoriasis, pointing to a causal role for hepcidin in the pathogenesis of the disease. Altogether, our data suggest that hepcidin could be an actionable target for skin psoriasis treatment, in addition to current therapeutics, or targeted as maintenance therapy during remission to prevent recurrence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- EQ202103012630/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- PMT202206015641/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- DEQ20160334903/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- ANR-11-IDEX-0005-02/Agence Nationale de la Recherche (French National Research Agency)
- ANR-11-LABX-0051/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

