The neonatal Fc receptor (FcRn) is a pan-arterivirus receptor
- PMID: 39112502
- PMCID: PMC11306234
- DOI: 10.1038/s41467-024-51142-x
The neonatal Fc receptor (FcRn) is a pan-arterivirus receptor
Abstract
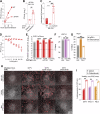
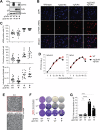
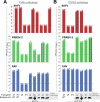
Arteriviruses infect a variety of mammalian hosts, but the receptors used by these viruses to enter cells are poorly understood. We identified the neonatal Fc receptor (FcRn) as an important pro-viral host factor via comparative genome-wide CRISPR-knockout screens with multiple arteriviruses. Using a panel of cell lines and divergent arteriviruses, we demonstrate that FcRn is required for the entry step of arterivirus infection and serves as a molecular barrier to arterivirus cross-species infection. We also show that FcRn synergizes with another known arterivirus entry factor, CD163, to mediate arterivirus entry. Overexpression of FcRn and CD163 sensitizes non-permissive cells to infection and enables the culture of fastidious arteriviruses. Treatment of multiple cell lines with a pre-clinical anti-FcRn monoclonal antibody blocked infection and rescued cells from arterivirus-induced death. Altogether, this study identifies FcRn as a novel pan-arterivirus receptor, with implications for arterivirus emergence, cross-species infection, and host-directed pan-arterivirus countermeasure development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Fields virology. (Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, PA, 2013).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

