ILC2-derived LIF licences progress from tissue to systemic immunity
- PMID: 39112698
- PMCID: PMC11338826
- DOI: 10.1038/s41586-024-07746-w
ILC2-derived LIF licences progress from tissue to systemic immunity
Abstract
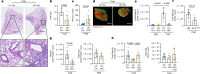
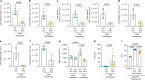
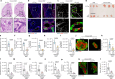
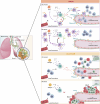
Migration and homing of immune cells are critical for immune surveillance. Trafficking is mediated by combinations of adhesion and chemokine receptors that guide immune cells, in response to chemokine signals, to specific locations within tissues and the lymphatic system to support tissue-localized immune reactions and systemic immunity1,2. Here we show that disruption of leukaemia inhibitory factor (LIF) production from group 2 innate lymphoid cells (ILC2s) prevents immune cells leaving the lungs to migrate to the lymph nodes (LNs). In the absence of LIF, viral infection leads to plasmacytoid dendritic cells (pDCs) becoming retained in the lungs where they improve tissue-localized, antiviral immunity, whereas chronic pulmonary allergen challenge leads to marked immune cell accumulation and the formation of tertiary lymphoid structures in the lung. In both cases immune cells fail to migrate to the lymphatics, leading to highly compromised LN reactions. Mechanistically, ILC2-derived LIF induces the production of the chemokine CCL21 from lymphatic endothelial cells lining the pulmonary lymphatic vessels, thus licensing the homing of CCR7+ immune cells (including dendritic cells) to LNs. Consequently, ILC2-derived LIF dictates the egress of immune cells from the lungs to regulate tissue-localized versus systemic immunity and the balance between allergen and viral responsiveness in the lungs.
© 2024. The Author(s).
Conflict of interest statement
A.N.J.M. is on the scientific advisory board of SinoMab. The other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

