Glycosphingolipid synthesis mediates immune evasion in KRAS-driven cancer
- PMID: 39112706
- PMCID: PMC11892728
- DOI: 10.1038/s41586-024-07787-1
Glycosphingolipid synthesis mediates immune evasion in KRAS-driven cancer
Abstract
Cancer cells frequently alter their lipids to grow and adapt to their environment1-3. Despite the critical functions of lipid metabolism in membrane physiology, signalling and energy production, how specific lipids contribute to tumorigenesis remains incompletely understood. Here, using functional genomics and lipidomic approaches, we identified de novo sphingolipid synthesis as an essential pathway for cancer immune evasion. Synthesis of sphingolipids is surprisingly dispensable for cancer cell proliferation in culture or in immunodeficient mice but required for tumour growth in multiple syngeneic models. Blocking sphingolipid production in cancer cells enhances the anti-proliferative effects of natural killer and CD8+ T cells partly via interferon-γ (IFNγ) signalling. Mechanistically, depletion of glycosphingolipids increases surface levels of IFNγ receptor subunit 1 (IFNGR1), which mediates IFNγ-induced growth arrest and pro-inflammatory signalling. Finally, pharmacological inhibition of glycosphingolipid synthesis synergizes with checkpoint blockade therapy to enhance anti-tumour immune response. Altogether, our work identifies glycosphingolipids as necessary and limiting metabolites for cancer immune evasion.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Declaration of Interests
K.B. is a scientific advisor to Nanocare Pharmaceuticals and Atavistik Bio. M.S. is an employee of Lime Therapeutics. The other authors have no competing interests.
Figures


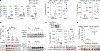
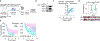





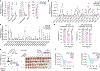




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

