Harnessing exosomes as cancer biomarkers in clinical oncology
- PMID: 39113040
- PMCID: PMC11308730
- DOI: 10.1186/s12935-024-03464-5
Harnessing exosomes as cancer biomarkers in clinical oncology
Abstract
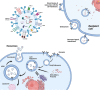
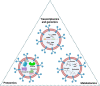
Exosomes are extracellular vesicles well known for facilitating cell-to-cell communication by distributing essential macromolecules like proteins, DNA, mRNA, lipids, and miRNA. These vesicles are abundant in fluids distributed throughout the body, including urine, blood, saliva, and even bile. They are important diagnostic tools for breast, lung, gastrointestinal cancers, etc. However, their application as cancer biomarkers has not yet been implemented in most parts of the world. In this review, we discuss how OMICs profiling of exosomes can be practiced by substituting traditional imaging or biopsy methods for cancer detection. Previous methods like extensive imaging and biopsy used for screening were expensive, mostly invasive, and could not easily provide early detection for various types of cancer. Exosomal biomarkers can be utilized for routine screening by simply collecting body fluids from the individual. We anticipate that the use of exosomes will be brought to light by the success of clinical trials investigating their potential to enhance cancer detection and treatment in the upcoming years.
Keywords: Cancer biomarkers; Cancer diagnosis; Clinical signature; Diagnostic tool; Exosomes; Molecular profiling; Prognostic indicator; Tumor-derived exosomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

