Neutrophils Recruited by NKX2-1 Suppression via Activation of CXCLs/CXCR2 Axis Promote Lung Adenocarcinoma Progression
- PMID: 39113226
- PMCID: PMC11481344
- DOI: 10.1002/advs.202400370
Neutrophils Recruited by NKX2-1 Suppression via Activation of CXCLs/CXCR2 Axis Promote Lung Adenocarcinoma Progression
Abstract
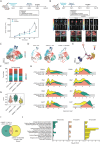
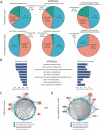
NK2 Homeobox 1 (NKX2-1) is a well-characterized pathological marker that delineates lung adenocarcinoma (LUAD) progression. The advancement of LUAD is influenced by the immune tumor microenvironment through paracrine signaling. However, the involvement of NKX2-1 in modeling the tumor immune microenvironment is still unclear. Here, the downregulation of NKX2-1 is observed in high-grade LUAD. Meanwhile, single-cell RNA sequencing and Visium in situ capturing profiling revealed the recruitment and infiltration of neutrophils in orthotopic syngeneic tumors exhibiting strong cell-cell communication through the activation of CXCLs/CXCR2 signaling. The depletion of NKX2-1 triggered the expression and secretion of CXCL1, CXCL2, CXCL3, and CXCL5 in LUAD cells. Chemokine secretion is analyzed by chemokine array and validated by qRT-PCR. ATAC-seq revealed the restrictive regulation of NKX2-1 on the promoters of CXCL1, CXCL2, and CXCL5 genes. This phenomenon led to increased tumor growth, and conversely, tumor growth decreased when inhibited by the CXCR2 antagonist SB225002. This study unveils how NKX2-1 modulates the infiltration of tumor-promoting neutrophils by inhibiting CXCLs/CXCR2-dependent mechanisms. Hence, targeting CXCR2 in NKX2-1-low tumors is a potential antitumor therapy that may improve LUAD patient outcomes.
Keywords: cheomkine; lung adenocarccinoma; single cell NGS; tumor microenvironment.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- La'ah A. S., Chiou S. H., Cells 2024, 13.
-
- Saito R. A., Watabe T., Horiguchi K., Kohyama T., Saitoh M., Nagase T., Miyazono K., Cancer Res. 2009, 69, 2783. - PubMed
MeSH terms
Substances
Grants and funding
- VGHUST111-G1-5-1/Taipei Veterans General Hospital
- VGHUST112-G1-5-1/Taipei Veterans General Hospital
- VGHUST113-G1-5-1/Taipei Veterans General Hospital
- VN111-15/Taipei Veterans General Hospital
- VN112-10/Taipei Veterans General Hospital
- V111C-157/Taipei Veterans General Hospital
- V111E-001-4/Taipei Veterans General Hospital
- V112E-002-4/Taipei Veterans General Hospital
- V112C-135/Taipei Veterans General Hospital
- V113C-064/Taipei Veterans General Hospital
- IDS2B/National Chiao Tung University
- 111VACS-008/Veterans Affairs Council, R.O.C.
- 112VACS-008/Veterans Affairs Council, R.O.C.
- IBMS-CRC109-P04/Institute of Biomedical Sciences, Academia Sinica
- IBMS-CRC112-P03/Institute of Biomedical Sciences, Academia Sinica
- MOST 109-2320-B-075-001/National Science and Technology Council
- MOST 110-2320-B-075-006-MY3/National Science and Technology Council
- NSTC112-2321-B-075-003/National Science and Technology Council
- NSTC113-2321-B-A49-013/National Science and Technology Council
LinkOut - more resources
Full Text Sources
Medical
Research Materials
