Loss of STK11 Suppresses Lipid Metabolism and Attenuates KRAS-Induced Immunogenicity in Patients with Non-Small Cell Lung Cancer
- PMID: 39113608
- PMCID: PMC11362717
- DOI: 10.1158/2767-9764.CRC-24-0153
Loss of STK11 Suppresses Lipid Metabolism and Attenuates KRAS-Induced Immunogenicity in Patients with Non-Small Cell Lung Cancer
Abstract
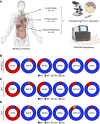
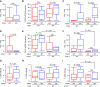
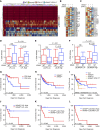
As many as 30% of the patients with non-small cell lung cancer harbor oncogenic KRAS mutations, which leads to extensive remodeling of the tumor immune microenvironment. Although co-mutations in several genes have prognostic relevance in KRAS-mutated patients, their effect on tumor immunogenicity are poorly understood. In the present study, a total of 189 patients with non-small cell lung cancer underwent a standardized analysis including IHC, whole-exome DNA sequencing, and whole-transcriptome RNA sequencing. Patients with activating KRAS mutations demonstrated a significant increase in PDL1 expression and CD8+ T-cell infiltration. Both were increased in the presence of a co-occurring TP53 mutation and lost with STK11 co-mutation. Subsequent genomic analysis demonstrated that KRAS/TP53 co-mutated tumors had a significant decrease in the expression of glycolysis-associated genes and an increase in several genes involved in lipid metabolism, notably lipoprotein lipase, low-density lipoprotein receptor, and LDLRAD4. Conversely, in the immune-excluded KRAS/STK11 co-mutated group, we observed diminished lipid metabolism and no change in anaerobic glycolysis. Interestingly, in patients with low expression of lipoprotein lipase, low-density lipoprotein receptor, or LDLRAD4, KRAS mutations had no effect on tumor immunogenicity. However, in patients with robust expression of these genes, KRAS mutations were associated with increased immunogenicity and associated with improved overall survival. Our data further suggest that the loss of STK11 may function as a metabolic switch, suppressing lipid metabolism in favor of glycolysis, thereby negating KRAS-induced immunogenicity. Hence, this concept warrants continued exploration, both as a predictive biomarker and potential target for therapy in patients receiving ICI-based immunotherapy.
Significance: In patients with lung cancer, we demonstrate that KRAS mutations increase tumor immunogenicity; however, KRAS/STK11 co-mutated patients display an immune-excluded phenotype. KRAS/STK11 co-mutated patients also demonstrated significant downregulation of several key lipid metabolism genes, many of which were associated with increased immunogenicity and improved overall survival in KRAS-mutated patients. Hence, alteration to lipid metabolism warrants further study as a potential biomarker and target for therapy in patients with KRAS-mutated lung cancer.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
R.H. Nguyen reports personal fees from Merck outside the submitted work. No disclosures were reported by the other authors.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

