Functional analysis of circSNYJ1/miR-142-5p/CCND1 regulatory axis in non-small cell lung cancer
- PMID: 39113852
- PMCID: PMC11301301
- DOI: 10.62347/PASE2970
Functional analysis of circSNYJ1/miR-142-5p/CCND1 regulatory axis in non-small cell lung cancer
Abstract
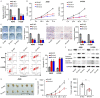
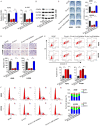
Non-small cell lung cancer (NSCLC), the most prevalent form of lung cancer, accounts for approximately 85% of all lung cancer diagnoses. Circular RNAs (circRNAs) are non-coding RNAs that play an active role in gene expression regulation, influencing cell growth, migration, and apoptosis. Here, we aimed to investigate the function of circSNYJ1 in NSCLC. In the present study, we found that circSNYJ1 expression level was increased in NSCLC tissues and cell lines. Knockdown of circSNYJ1 suppressed NSCLC cell proliferation, colony formation and migration while promoting apoptosis. Mechanistically, we demonstrated that circSNYJ1 sponged miR-142-5p, thereby regulating the expression of CCND1, a well-known cell cycle regulator. In conclusion, this study uncovered a novel circSNYJ1/miR-142-5p/CCND1 axis involved in NSCLC progression, providing potential diagnostic and prognostic biomarkers for treating NSCLC.
Keywords: CCND1; CircSNYJ1; NSCLC; biomarkers; miR-142-5p.
AJCR Copyright © 2024.
Conflict of interest statement
None.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
