The Spatial Structure of the Tumor Immune Microenvironment Can Explain and Predict Patient Response in High-Grade Serous Carcinoma
- PMID: 39115368
- PMCID: PMC11534564
- DOI: 10.1158/2326-6066.CIR-23-1109
The Spatial Structure of the Tumor Immune Microenvironment Can Explain and Predict Patient Response in High-Grade Serous Carcinoma
Abstract
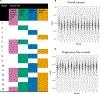
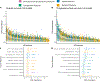
Ovarian cancer is the deadliest gynecologic malignancy, and therapeutic options and mortality rates over the last three decades have largely not changed. Recent studies indicate that the composition of the tumor immune microenvironment (TIME) influences patient outcomes. To improve spatial understanding of the TIME, we performed multiplexed ion beam imaging on 83 human high-grade serous carcinoma tumor samples, identifying approximately 160,000 cells across 23 cell types. From the 77 of these samples that met inclusion criteria, we generated composition features based on cell type proportions, spatial features based on the distances between cell types, and spatial network features representing cell interactions and cell clustering patterns, which we linked to traditional clinical and IHC variables and patient overall survival (OS) and progression-free survival (PFS) outcomes. Among these features, we found several significant univariate correlations, including B-cell contact with M1 macrophages (OS HR = 0.696; P = 0.011; PFS HR = 0.734; P = 0.039). We then used high-dimensional random forest models to evaluate out-of-sample predictive performance for OS and PFS outcomes and to derive relative feature importance scores for each feature. The top model for predicting low or high PFS used TIME composition and spatial features and achieved an average AUC score of 0.71. The results demonstrate the importance of spatial structure in understanding how the TIME contributes to treatment outcomes. Furthermore, the present study provides a generalizable roadmap for spatial analyses of the TIME in ovarian cancer research.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures





Update of
-
The spatial structure of the tumor immune microenvironment can explain and predict patient response in high-grade serous carcinoma.bioRxiv [Preprint]. 2024 Jan 29:2024.01.26.577350. doi: 10.1101/2024.01.26.577350. bioRxiv. 2024. Update in: Cancer Immunol Res. 2024 Nov 4;12(11):1492-1507. doi: 10.1158/2326-6066.CIR-23-1109. PMID: 38352574 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

