AMPK activation induces RALDH+ tolerogenic dendritic cells by rewiring glucose and lipid metabolism
- PMID: 39115541
- PMCID: PMC11310580
- DOI: 10.1083/jcb.202401024
AMPK activation induces RALDH+ tolerogenic dendritic cells by rewiring glucose and lipid metabolism
Abstract
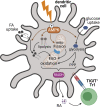
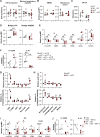
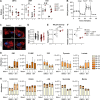
Dendritic cell (DC) activation and function are underpinned by profound changes in cellular metabolism. Several studies indicate that the ability of DCs to promote tolerance is dependent on catabolic metabolism. Yet the contribution of AMP-activated kinase (AMPK), a central energy sensor promoting catabolism, to DC tolerogenicity remains unknown. Here, we show that AMPK activation renders human monocyte-derived DCs tolerogenic as evidenced by an enhanced ability to drive differentiation of regulatory T cells, a process dependent on increased RALDH activity. This is accompanied by several metabolic changes, including increased breakdown of glycerophospholipids, enhanced mitochondrial fission-dependent fatty acid oxidation, and upregulated glucose catabolism. This metabolic rewiring is functionally important as we found interference with these metabolic processes to reduce to various degrees AMPK-induced RALDH activity as well as the tolerogenic capacity of moDCs. Altogether, our findings reveal a key role for AMPK signaling in shaping DC tolerogenicity and suggest AMPK as a target to direct DC-driven tolerogenic responses in therapeutic settings.
© 2024 Brombacher et al.
Conflict of interest statement
Disclosures: C.R. Berkers reported grants from Trajectum Pharma outside the submitted work. No other disclosures were reported.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

